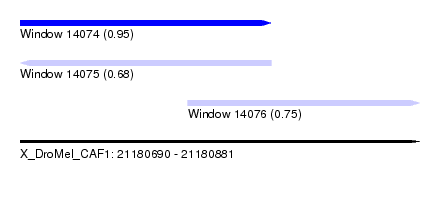
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 21,180,690 – 21,180,881 |
| Length | 191 |
| Max. P | 0.947933 |

| Location | 21,180,690 – 21,180,810 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.67 |
| Mean single sequence MFE | -39.98 |
| Consensus MFE | -33.12 |
| Energy contribution | -33.50 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.947933 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21180690 120 + 22224390 AAGAAGCCUUGAGGGAUAUUCUCCGGCAGCGGCAUCUUACGUUUCGGGCUUGUCCAGGGACUUUUUGAUAUCCAGAGAACUUUCCCAAAGAUCAGGAGAAUUCCUGACAGCUGCUGCAAA .((((.((.....))...))))...((((((((.((((.......(((....))).((((.((((((.....))))))....)))).))))(((((......)))))..))))))))... ( -39.80) >DroSec_CAF1 6879 120 + 1 AAAAAGCCUUGUGGGAUAUUCUCCGGCAGCGGCAUCUUACGUAUCAGGCUUGUCCAGGGACAUUUUGAUAUCCAGAGAACUUUCCCAAAGAUCAGGAGACUUCCUGACAGCUGCUGCAAA .............(((.....))).((((((((.((((..((((((((..((((....)))).))))))))...)))).............(((((......)))))..))))))))... ( -39.20) >DroEre_CAF1 7006 120 + 1 AGGAAGCCUUGUGGGAUUUUCUCCGGCAACGGCAUCCUAUGUUUCAGGCUUGUCCAGGGAUUUCUUGAUAUCCAGAGAACUUUCCCAAAGAUCGGGAGAUUUCCUGACAGCUGCUGCAAA ((((.(((((((.(((.....))).)))).))).)))).(((....((((.(((.((((((((((.(....).)))))..((((((.......)))))).))))))))))))...))).. ( -43.50) >DroYak_CAF1 215 120 + 1 AGGAAGCCUUGUGGGAUUUUCUCCGGCAACAGCAUCCUAUGUUUCAGGCUUGUCCAGGGAUUUCUUGAUAUCCAGAGAACUAUCCCAAAGAUCAGGAGAUUUUCUGACAGCUGCUGCAAA ((((.((.((((.(((.....))).))))..)).)))).(((....((((.(((..(((((((((((((.....((......))......)))))))))))))..)))))))...))).. ( -37.40) >consensus AAGAAGCCUUGUGGGAUAUUCUCCGGCAACGGCAUCCUACGUUUCAGGCUUGUCCAGGGACUUCUUGAUAUCCAGAGAACUUUCCCAAAGAUCAGGAGAUUUCCUGACAGCUGCUGCAAA ((((.(((((((.(((.....))).)))).))).))))..((....((((.(((.((((((((((((((.....(((....)))......))))))))).))))))))))))...))... (-33.12 = -33.50 + 0.38)

| Location | 21,180,690 – 21,180,810 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.67 |
| Mean single sequence MFE | -35.88 |
| Consensus MFE | -30.69 |
| Energy contribution | -30.75 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.683003 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21180690 120 - 22224390 UUUGCAGCAGCUGUCAGGAAUUCUCCUGAUCUUUGGGAAAGUUCUCUGGAUAUCAAAAAGUCCCUGGACAAGCCCGAAACGUAAGAUGCCGCUGCCGGAGAAUAUCCCUCAAGGCUUCUU ...(((((.((.(((((((....))))))).((((((...((((...((((........))))..))))...)))))).........)).))))).(((.....)))............. ( -37.20) >DroSec_CAF1 6879 120 - 1 UUUGCAGCAGCUGUCAGGAAGUCUCCUGAUCUUUGGGAAAGUUCUCUGGAUAUCAAAAUGUCCCUGGACAAGCCUGAUACGUAAGAUGCCGCUGCCGGAGAAUAUCCCACAAGGCUUUUU .....(((....(((((((....)))))))((((((((..(((((((((.(((((...((((....))))....)))))(....).........))))))))).))))).)))))).... ( -39.60) >DroEre_CAF1 7006 120 - 1 UUUGCAGCAGCUGUCAGGAAAUCUCCCGAUCUUUGGGAAAGUUCUCUGGAUAUCAAGAAAUCCCUGGACAAGCCUGAAACAUAGGAUGCCGUUGCCGGAGAAAAUCCCACAAGGCUUCCU ...((....))..(((((.....((((((...))))))..((((...((((........))))..))))...))))).....((((.(((.(((..(((.....)))..)))))).)))) ( -34.80) >DroYak_CAF1 215 120 - 1 UUUGCAGCAGCUGUCAGAAAAUCUCCUGAUCUUUGGGAUAGUUCUCUGGAUAUCAAGAAAUCCCUGGACAAGCCUGAAACAUAGGAUGCUGUUGCCGGAGAAAAUCCCACAAGGCUUCCU ...((((((((.(((((........))))).((..((...((((...((((........))))..))))...))..)).........)))))))).(((.....)))............. ( -31.90) >consensus UUUGCAGCAGCUGUCAGGAAAUCUCCUGAUCUUUGGGAAAGUUCUCUGGAUAUCAAAAAAUCCCUGGACAAGCCUGAAACAUAAGAUGCCGCUGCCGGAGAAAAUCCCACAAGGCUUCCU ...(((((.((.(((((((....))))))).((((((...((((...((((........))))..))))...)))))).........)).))))).(((.....)))............. (-30.69 = -30.75 + 0.06)


| Location | 21,180,770 – 21,180,881 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 93.09 |
| Mean single sequence MFE | -32.03 |
| Consensus MFE | -28.46 |
| Energy contribution | -28.27 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.749210 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21180770 111 + 22224390 UUUCCCAAAGAUCAGGAGAAUUCCUGACAGCUGCUGCAAAGAUGGUAUCCCAAAUGGGAGCAAGCGAACUCCGCUGGCUCCCCAUAUCCAGGACCAAAGACAAACCAUCGA ...........(((((......)))))..((....))...((((((.(((.....((((((.((((.....)))).))))))........)))(....)....)))))).. ( -33.42) >DroSec_CAF1 6959 111 + 1 UUUCCCAAAGAUCAGGAGACUUCCUGACAGCUGCUGCAAAGAUGGUAUCCCAAAUGGGAGCAAGCGAACUCCGCUGGUUGCCCAUACCCAGGACCAAAGACAAACCAUCGA ...........(((((......)))))..((....))...((((((.......((((((((.((((.....)))).))).))))).(....)...........)))))).. ( -31.10) >DroEre_CAF1 7086 111 + 1 UUUCCCAAAGAUCGGGAGAUUUCCUGACAGCUGCUGCAAAAAUGGUAUCCCAAAUGGGAGCAGGCGAACUCCGCUGGAUGCCAAUGCCCAGGACCAAAGAUAAGCCAUCGA ((((((.......))))))..(((((...((....)).....((((((((....((....))((((.....)))))))))))).....))))).....(((.....))).. ( -32.10) >DroYak_CAF1 295 111 + 1 UAUCCCAAAGAUCAGGAGAUUUUCUGACAGCUGCUGCAAAGAUGGUAUCCCAAAUGGGAGCAAGCGAACUCCGCUGGCUGCCCAUGCCCAGGACCAAAGAUAAGCCAUCGA ...........((((((....))))))..((....))...((((((.(((...((((((((.((((.....)))).))).))))).....)))(....)....)))))).. ( -31.50) >consensus UUUCCCAAAGAUCAGGAGAUUUCCUGACAGCUGCUGCAAAGAUGGUAUCCCAAAUGGGAGCAAGCGAACUCCGCUGGCUGCCCAUACCCAGGACCAAAGACAAACCAUCGA ...........((((((....))))))..((....))...((((((.(((...((((((((.((((.....)))).))).))))).....)))(....)....)))))).. (-28.46 = -28.27 + -0.19)


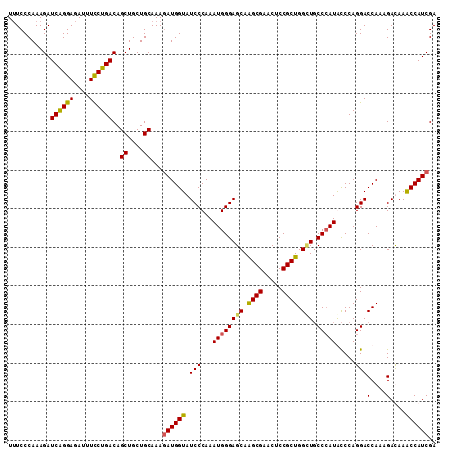
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:07:10 2006