
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 21,161,237 – 21,161,397 |
| Length | 160 |
| Max. P | 0.898655 |

| Location | 21,161,237 – 21,161,357 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.33 |
| Mean single sequence MFE | -36.62 |
| Consensus MFE | -30.78 |
| Energy contribution | -31.46 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.867977 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
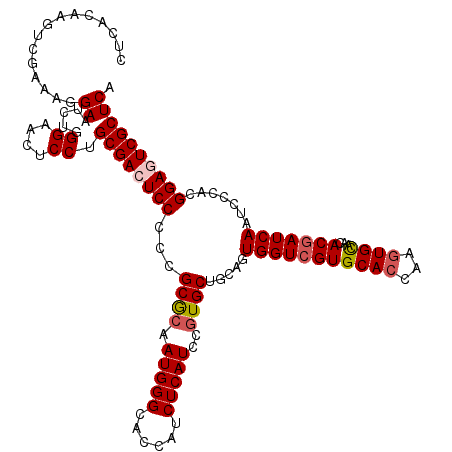
>X_DroMel_CAF1 21161237 120 + 22224390 CUCAGAAGUCGAAAGGAAUCUGGGAACUCCUGCGACUCCCCCGCGCAAUGGGCACCAUCUCAUCCGUGCUGCAGUGGUCGUGCACCAAAUGUAACACUAUCAAUCCCACGGAGUCGCUCA ((((((..((....))..)))))).......((((((((...((((.(((((......)))))..))))...((((....((((.....)))).))))...........))))))))... ( -40.20) >DroSec_CAF1 5182 120 + 1 CUCACAAGUCGAAAGGAAUCUGGGAACUCCUGCGACUCCCCCGCGCAAUGGGCACCAUCUCAUCCGUGCUGCAGUGGUCGUGCACCAAGUGCAACACGAUCAAUCCCACGGAGUCGCUCA ((((....((....))....)))).......((((((((...((((.(((((......)))))..)))).....(((((((((((...))))...))))))).......))))))))... ( -40.20) >DroSim_CAF1 5131 120 + 1 CUCACAAGUCGAAAGGAAUCUGGGAACUCCUGCGACUCCCCCGCGCAAUGGGCACCAUCUCAUCCGUGCUGCAGUGGUCGUGCACCAAGUGCAACACGAUCAAUCCCACGGAGUCGCUCA ((((....((....))....)))).......((((((((...((((.(((((......)))))..)))).....(((((((((((...))))...))))))).......))))))))... ( -40.20) >DroEre_CAF1 5906 120 + 1 CUCACAACUCGAAACGAAUCUCGGAACUCCUGCGACUCCCCCGCGAAAUGGGCACCAUCUCAUCCGUGCUGCAGUGGUCGUGCACCAAGUGCAACACGAUCAAUCCCACGGAAUCGCUCA ..........((...((.((.(((.....))).)).))....((((..((((......))))((((((......(((((((((((...))))...)))))))....)))))).)))))). ( -30.90) >DroYak_CAF1 5259 120 + 1 CUCACAACUCUAAACGAAUCUCGGAACUCCUGCGACUCCCCUGCACAAUGGGCACCAUCUCAUCCGUGCUGCAGUGGUCGUGCACCAAGUGCAACACGAUCAAUCCCACGGAAUCGCUCA ...............((.....((....)).((((.(((.(((((..(((((..........)))))..)))))(((((((((((...))))...))))))).......))).)))))). ( -31.60) >consensus CUCACAAGUCGAAAGGAAUCUGGGAACUCCUGCGACUCCCCCGCGCAAUGGGCACCAUCUCAUCCGUGCUGCAGUGGUCGUGCACCAAGUGCAACACGAUCAAUCCCACGGAGUCGCUCA ...............((.....((....)).((((((((...((((.(((((......)))))..)))).....(((((((((((...))))...))))))).......)))))))))). (-30.78 = -31.46 + 0.68)

| Location | 21,161,277 – 21,161,397 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -39.63 |
| Consensus MFE | -37.40 |
| Energy contribution | -37.24 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.898655 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
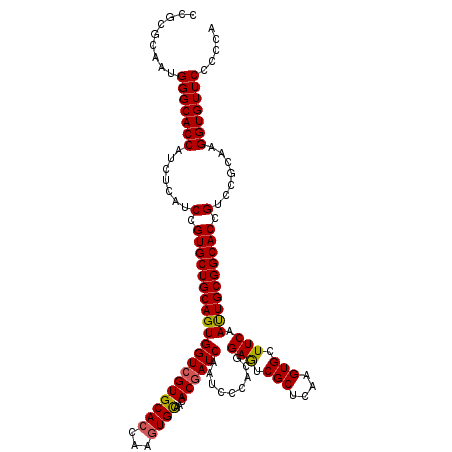
>X_DroMel_CAF1 21161277 120 + 22224390 CCGCGCAAUGGGCACCAUCUCAUCCGUGCUGCAGUGGUCGUGCACCAAAUGUAACACUAUCAAUCCCACGGAGUCGCUCAAGUGCUUCAAUUGCGGCACCGUCCGCAAGGUGUUCCCCCA ...((((((.(((((.......((((((.....(((((..((((.....))))..)))))......)))))).........)))))...))))))(((((........)))))....... ( -34.19) >DroSec_CAF1 5222 120 + 1 CCGCGCAAUGGGCACCAUCUCAUCCGUGCUGCAGUGGUCGUGCACCAAGUGCAACACGAUCAAUCCCACGGAGUCGCUCAAGUGCUUCAAUUGCGGCACUGUCCGCAAGGUGUUCCCCCA .........(((((((.........((((((((((((((((((((...))))...))))))........((((.(((....))))))).))))))))))((....)).)))))))..... ( -41.80) >DroSim_CAF1 5171 120 + 1 CCGCGCAAUGGGCACCAUCUCAUCCGUGCUGCAGUGGUCGUGCACCAAGUGCAACACGAUCAAUCCCACGGAGUCGCUCAAGUGCUUCAAUUGCGGCACUGUCCGCAAGGUGUUCCCCCA .........(((((((.........((((((((((((((((((((...))))...))))))........((((.(((....))))))).))))))))))((....)).)))))))..... ( -41.80) >DroEre_CAF1 5946 120 + 1 CCGCGAAAUGGGCACCAUCUCAUCCGUGCUGCAGUGGUCGUGCACCAAGUGCAACACGAUCAAUCCCACGGAAUCGCUCAAGUGCUUCAACUGCGGCACCGUCCGCAAGGUGUUCCCCCA .........(((((((.......(.((((((((((((((((((((...))))...)))))).........(((.(((....))).))).)))))))))).).......)))))))..... ( -41.04) >DroYak_CAF1 5299 120 + 1 CUGCACAAUGGGCACCAUCUCAUCCGUGCUGCAGUGGUCGUGCACCAAGUGCAACACGAUCAAUCCCACGGAAUCGCUCAAGUGCUUCAAUUGCGGCACCGUCCGCAAGGUGUUCCCCCA .........(((((((......((((((......(((((((((((...))))...)))))))....))))))...((......)).....((((((......)))))))))))))..... ( -39.30) >consensus CCGCGCAAUGGGCACCAUCUCAUCCGUGCUGCAGUGGUCGUGCACCAAGUGCAACACGAUCAAUCCCACGGAGUCGCUCAAGUGCUUCAAUUGCGGCACCGUCCGCAAGGUGUUCCCCCA .........(((((((.......(.((((((((((((((((((((...))))...)))))).........(((.(((....))).))).)))))))))).).......)))))))..... (-37.40 = -37.24 + -0.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:06:56 2006