| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 2,426,079 – 2,426,170 |
| Length | 91 |
| Max. P | 0.992986 |

| Location | 2,426,079 – 2,426,170 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 72.67 |
| Mean single sequence MFE | -23.14 |
| Consensus MFE | -9.39 |
| Energy contribution | -10.20 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.41 |
| SVM decision value | 2.37 |
| SVM RNA-class probability | 0.992986 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
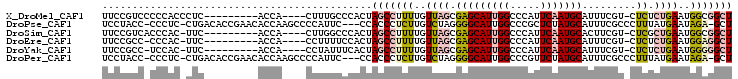
>X_DroMel_CAF1 2426079 91 + 22224390 UUCCGUCCCCCACCCUC---------ACCA----CUUUGCCCACUAGCCUUUUGUUAGCGAGCAUUGGCCCAUUCAAUGCAUUUCGU-CUCUCUGAAUGGCGGCU ..(((............---------.(((----...(((.(.(((((.....))))).).))).))).(((((((...........-.....)))))))))).. ( -18.69) >DroPse_CAF1 103083 99 + 1 UCCUACC-CCCUC-CUGACACCGAACACCAAGCCCCAUUC---CCACCCUCUUGUCUAGGGGCAUUGGCCCGCUCUAUGCAUUUCGCCCUUUAUGAAUAGA-GCU .......-.....-.............(((((((((....---...............))))).))))...(((((((.(((..........))).)))))-)). ( -23.41) >DroSim_CAF1 35817 90 + 1 UUCCGUCACCCAC-UUC---------ACCA----CUUGGCCCACUAGCCUUUUGUUAGCGAGCAUUGGCCCAUUCAAUGCACUUCGU-CUCGCUGAAUGGCGGCU ..((((((.....-...---------....----...(((......))).....((((((((((((((.....))))))).......-.))))))).)))))).. ( -25.10) >DroEre_CAF1 71951 89 + 1 UUCCGCC-CCCAC-UUC---------ACCA----CCUUUUCCACUAGCCUUUUGUUAGCGAGCAUUGGCCCAUUCAAUGCAUUUCGU-CUCUCUGAAUGGAGGCU ....(((-.(((.-(((---------(...----.......(.(((((.....))))).).(((((((.....))))))).......-.....))))))).))). ( -22.60) >DroYak_CAF1 72913 89 + 1 UUCCGCC-UCCAC-UUC---------ACCA----CCUAUUUCACUAGCCUUUUGUUAGCGAGCAUUGGCCCAUUCAAUGCAUUUCGU-CUCUCUGAAUGGGGGCU ....(((-((((.-(((---------(...----......((.(((((.....))))).))(((((((.....))))))).......-.....))))))))))). ( -28.00) >DroPer_CAF1 103878 99 + 1 UCCUACC-CCCUC-CUGACACCGAACACCAAGCCCCAUUC---CCACCCUCUUGUCUAGGGGCAUUGGCCCGUUCUAUGCAUUUCGCCCUUUAUGAAUAGA-GCU .......-.....-.............(((((((((....---...............))))).))))...(((((((.(((..........))).)))))-)). ( -21.01) >consensus UUCCGCC_CCCAC_CUC_________ACCA____CCUUUCCCACUAGCCUUUUGUUAGCGAGCAUUGGCCCAUUCAAUGCAUUUCGU_CUCUCUGAAUGGAGGCU .............................................(((((((..((((.(((((((((.....))))))).........)).))))..))))))) ( -9.39 = -10.20 + 0.81)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:55:20 2006