
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 21,155,879 – 21,156,063 |
| Length | 184 |
| Max. P | 0.951305 |

| Location | 21,155,879 – 21,155,984 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 84.13 |
| Mean single sequence MFE | -32.13 |
| Consensus MFE | -26.59 |
| Energy contribution | -26.60 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.628909 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21155879 105 + 22224390 ACAUUAUUGUUAGUUCGCUGCUAUUAUAUUGGCCUCAGCUGUAUCUGCGGUUAGCGAGUGCACCCACCGCAGUUAGCACGUGCCAGCACCUUAUUUGGCGAAUUC ...........((((((((((((......))))....((((...(((((((..((....))....))))))).))))..(((....))).......)))))))). ( -30.70) >DroSec_CAF1 20058 105 + 1 ACAUUUUAGUUAGUUCGCUGCUAUUAUUUUGGCCCCAGCUGUAGCUGCGGUUAGCGAGUGCACCCACCGCAGUUAGCACGUGCCAGCACCGCAUUUGGCGAACUC ...........((((((((..........(((((.((((....)))).)))))(((.((((...(((.((.....))..)))...)))))))....)))))))). ( -36.40) >DroYak_CAF1 20778 104 + 1 UCAUUUUUGCUAUUUCGCUGCUAUUAUUUUGGCCUUAGCGGUACCCAC-GUUAGCGAGUGCACACACCGCAGUAAGCGAGUGUCCGCACCACAUUUGGCCAGUUC ........((((..((((.((((......))))..(((((.......)-))))))))((((..(((((((.....))).))))..))))......))))...... ( -29.30) >consensus ACAUUUUUGUUAGUUCGCUGCUAUUAUUUUGGCCUCAGCUGUACCUGCGGUUAGCGAGUGCACCCACCGCAGUUAGCACGUGCCAGCACCACAUUUGGCGAAUUC ...........((((((((((((......))))....(((((((.(((.....(((.(((....)))))).....))).))).)))).........)))))))). (-26.59 = -26.60 + 0.01)

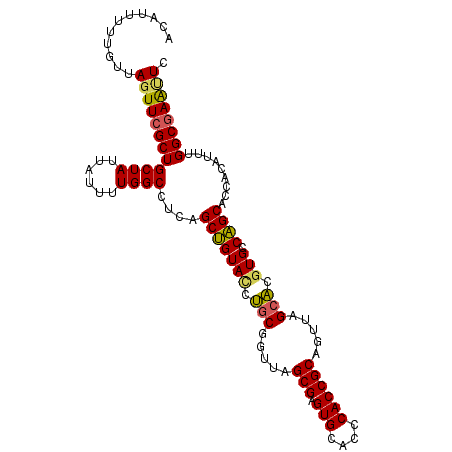

| Location | 21,155,879 – 21,155,984 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 84.13 |
| Mean single sequence MFE | -37.10 |
| Consensus MFE | -21.64 |
| Energy contribution | -22.53 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.86 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.693907 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21155879 105 - 22224390 GAAUUCGCCAAAUAAGGUGCUGGCACGUGCUAACUGCGGUGGGUGCACUCGCUAACCGCAGAUACAGCUGAGGCCAAUAUAAUAGCAGCGAACUAACAAUAAUGU ...(((((....((..(((.((((....(((..(((((((.((((....)))).)))))))....)))....)))).)))..))...)))))............. ( -32.20) >DroSec_CAF1 20058 105 - 1 GAGUUCGCCAAAUGCGGUGCUGGCACGUGCUAACUGCGGUGGGUGCACUCGCUAACCGCAGCUACAGCUGGGGCCAAAAUAAUAGCAGCGAACUAACUAAAAUGU .(((((((....(((.((..((((....(((..(((((((.((((....)))).)))))))....)))....))))..))....))))))))))........... ( -38.20) >DroYak_CAF1 20778 104 - 1 GAACUGGCCAAAUGUGGUGCGGACACUCGCUUACUGCGGUGUGUGCACUCGCUAAC-GUGGGUACCGCUAAGGCCAAAAUAAUAGCAGCGAAAUAGCAAAAAUGA ....(((((....((((((((.((((.(((.....))))))).)))((((((....-)))))))))))...)))))...........((......))........ ( -40.90) >consensus GAAUUCGCCAAAUGAGGUGCUGGCACGUGCUAACUGCGGUGGGUGCACUCGCUAACCGCAGAUACAGCUGAGGCCAAAAUAAUAGCAGCGAACUAACAAAAAUGU ....((((........(((....)))..((((.(((((((.((((....)))).))))))).....((....))........)))).)))).............. (-21.64 = -22.53 + 0.89)

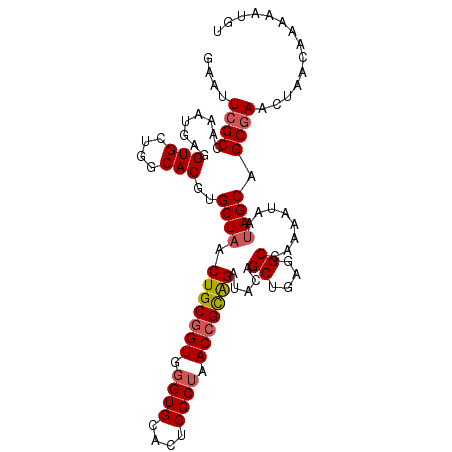

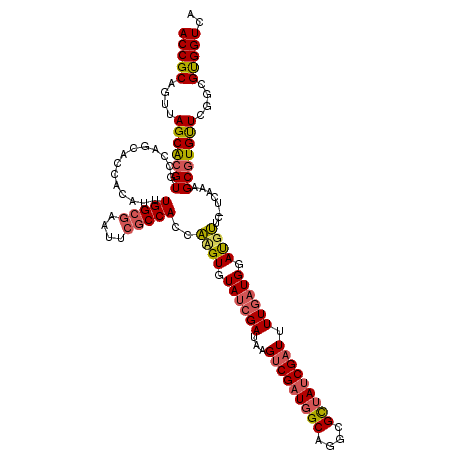
| Location | 21,155,904 – 21,156,024 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.22 |
| Mean single sequence MFE | -49.03 |
| Consensus MFE | -33.18 |
| Energy contribution | -33.63 |
| Covariance contribution | 0.46 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.951305 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21155904 120 - 22224390 CGAUAGCGCCUGCCAUCGACAUAUCGAUACACAUGGUGGCGAAUUCGCCAAAUAAGGUGCUGGCACGUGCUAACUGCGGUGGGUGCACUCGCUAACCGCAGAUACAGCUGAGGCCAAUAU ....(((((((((((((((....))))......))))((((....)))).....)))))))(((....(((..(((((((.((((....)))).)))))))....)))....)))..... ( -47.40) >DroSec_CAF1 20083 120 - 1 CGAUAGCGCCUGCCAUCGACGUAUCGAUACACUUGGUGGCGAGUUCGCCAAAUGCGGUGCUGGCACGUGCUAACUGCGGUGGGUGCACUCGCUAACCGCAGCUACAGCUGGGGCCAAAAU ...((((....((((((((.((........))))))))))((((.((((...((((((..((((....))))))))))...)))).)))))))).((.((((....)))).))....... ( -49.40) >DroYak_CAF1 20803 118 - 1 CGAUAACGCCUGCGA-CGGCUUAUCGGUAUACGAUGUGGUGAACUGGCCAAAUGUGGUGCGGACACUCGCUUACUGCGGUGUGUGCACUCGCUAAC-GUGGGUACCGCUAAGGCCAAAAU ((((((.(((.....-.)))))))))((.(((......))).))(((((....((((((((.((((.(((.....))))))).)))((((((....-)))))))))))...))))).... ( -50.30) >consensus CGAUAGCGCCUGCCAUCGACAUAUCGAUACACAUGGUGGCGAAUUCGCCAAAUGAGGUGCUGGCACGUGCUAACUGCGGUGGGUGCACUCGCUAACCGCAGAUACAGCUGAGGCCAAAAU .......((((((((((((....)))))........(((((....))))).....)))((((((....))...(((((((.((((....)))).)))))))...))))..))))...... (-33.18 = -33.63 + 0.46)

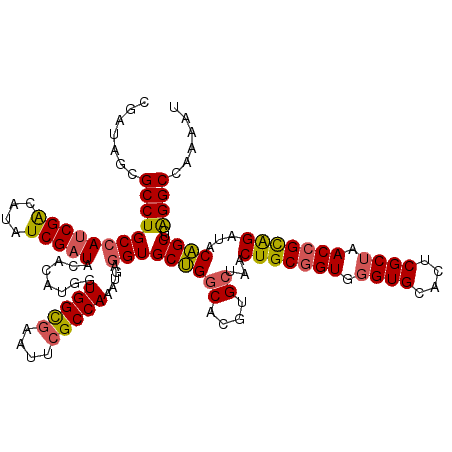
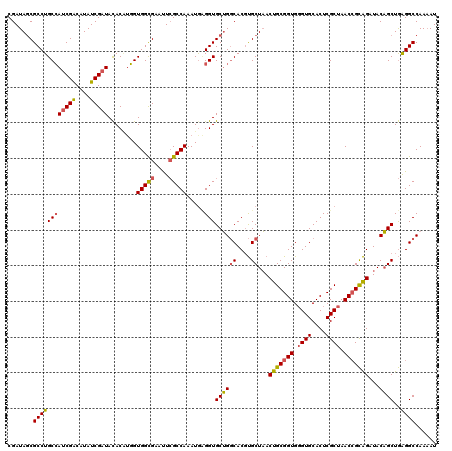
| Location | 21,155,944 – 21,156,063 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.89 |
| Mean single sequence MFE | -43.53 |
| Consensus MFE | -31.99 |
| Energy contribution | -33.33 |
| Covariance contribution | 1.35 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.695325 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21155944 119 + 22224390 ACCGCAGUUAGCACGUGCCAGCACCUUAUUUGGCGAAUUCGCCACCAUGUGUAUCGAUAUGUCGAUGGCAGGCGCUAUCGAUUUUGAUGGAUGUUC-UCAAAGCGUGUUCGGCGCGGUCA (((((.....))..(((((((((((((...(((((....)))))..((((.((((((...(((((((((....))))))))).)))))).))))..-...))).))))).)))))))).. ( -47.80) >DroSec_CAF1 20123 119 + 1 ACCGCAGUUAGCACGUGCCAGCACCGCAUUUGGCGAACUCGCCACCAAGUGUAUCGAUACGUCGAUGGCAGGCGCUAUCGAUUUUGAUGGAUGUUC-UCAAAGCGUGCUCGGGGUGGUCA (((((....(((((((...((((((((((((((((....)))))...)))))(((((...(((((((((....))))))))).))))))).)))).-.....)))))))....))))).. ( -47.50) >DroYak_CAF1 20842 119 + 1 ACCGCAGUAAGCGAGUGUCCGCACCACAUUUGGCCAGUUCACCACAUCGUAUACCGAUAAGCCG-UCGCAGGCGUUAUCGAUUUUUAUGGACGGUAUUGAUAGCGUGUUCGGCGUGGUCA (((((......(((((((.......)))))))(((.(..(((...((((.((((((....(((.-.....)))....((.((....)).))))))))))))...)))..))))))))).. ( -35.30) >consensus ACCGCAGUUAGCACGUGCCAGCACCACAUUUGGCGAAUUCGCCACCAAGUGUAUCGAUAAGUCGAUGGCAGGCGCUAUCGAUUUUGAUGGAUGUUC_UCAAAGCGUGUUCGGCGUGGUCA (((((....(((((((..............(((((....)))))..((((.((((((...(((((((((....))))))))).)))))).))))........)))))))....))))).. (-31.99 = -33.33 + 1.35)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:06:51 2006