| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 21,126,585 – 21,126,696 |
| Length | 111 |
| Max. P | 0.993534 |

| Location | 21,126,585 – 21,126,696 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 68.45 |
| Mean single sequence MFE | -21.06 |
| Consensus MFE | -9.65 |
| Energy contribution | -11.49 |
| Covariance contribution | 1.84 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.46 |
| SVM decision value | 2.41 |
| SVM RNA-class probability | 0.993534 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21126585 111 + 22224390 GGUCGUGUUUUGUUAUAUUUAAUUUUAAUUGUUUACUGGACACAUUUA-------AUUC-GAAAACCUUAGCUAGGUUUUUUACCACCUAGCGUUU-UUAUCGAUGCCGAUUGCUCGAUG ....(((((..((.(((.(((....))).)))..))..))))).....-------....-((((((....(((((((........)))))))))))-))(((((.((.....))))))). ( -24.80) >DroSec_CAF1 7712 108 + 1 CGCCGAGUUUUAUUACAUUUACUUUUAAUUGGUUACUUGGCACACUUA-------AUUC-GAAAACCUUAGCUAGGCUC---CCCACCUAGCGUUU-UUAUCGAUACCGAUUACUCAAUU .(((((((.......((.(((....))).))...))))))).......-------....-((((((....((((((...---....))))))))))-))((((....))))......... ( -22.10) >DroSim_CAF1 7617 108 + 1 CGCCGAGUUUUCUUACAUUUACUUUUAAUUGGUUACUUGGCACACUUA-------AUUC-GAAAUCCUUAGCUAGGCUC---CCCACCUAGCGUUU-UUAUCGAUACCGAUUACUCGAUU .(((((((..((..................))..))))))).......-------..((-((........((((((...---....))))))((..-..((((....)))).)))))).. ( -26.07) >DroEre_CAF1 7116 110 + 1 ----GAAU--UUUUUAAUGUACUUUUAAGCUGCUUUUUUAAGCAAUUAAAUUUCAAUUUAAAAAACCUUAGCUAGGUUU---CCCACCUAUCCAUU-UUAUCGAUUCCGAUUACUCUGUG ----...(--(((((((......(((((..(((((....))))).))))).......)))))))).......(((((..---...)))))..(((.-..((((....))))......))) ( -14.62) >DroYak_CAF1 8051 86 + 1 ----AUAU--CUUCUA-----C------------AUUUGAAACAUUUA-------AUUU-GAGAACAUUAGCUAGGUUU---CCCACCUAGCUAUUCUUAUCGAUUCCGAUUACUCUGUG ----....--....((-----(------------(.............-------...(-(((((...(((((((((..---...)))))))))))))))(((....)))......)))) ( -17.70) >consensus _G_CGAGUUUUUUUAAAUUUACUUUUAAUUGGUUACUUGACACACUUA_______AUUC_GAAAACCUUAGCUAGGUUU___CCCACCUAGCGUUU_UUAUCGAUACCGAUUACUCGAUG .(((((((..........................))))))).............................(((((((........))))))).......((((....))))......... ( -9.65 = -11.49 + 1.84)

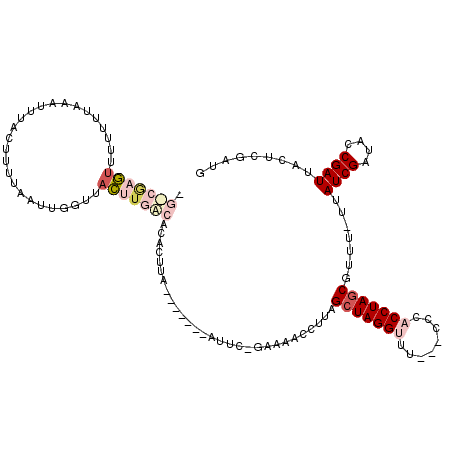
| Location | 21,126,585 – 21,126,696 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 68.45 |
| Mean single sequence MFE | -20.76 |
| Consensus MFE | -13.12 |
| Energy contribution | -13.92 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.885562 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21126585 111 - 22224390 CAUCGAGCAAUCGGCAUCGAUAA-AAACGCUAGGUGGUAAAAAACCUAGCUAAGGUUUUC-GAAU-------UAAAUGUGUCCAGUAAACAAUUAAAAUUAAAUAUAACAAAACACGACC ..((((....))))..((((.((-....(((((((........))))))).....)).))-)).(-------(((.(((.........))).))))........................ ( -19.80) >DroSec_CAF1 7712 108 - 1 AAUUGAGUAAUCGGUAUCGAUAA-AAACGCUAGGUGGG---GAGCCUAGCUAAGGUUUUC-GAAU-------UAAGUGUGCCAAGUAACCAAUUAAAAGUAAAUGUAAUAAAACUCGGCG .(((((....))))).((((.((-....(((((((...---..))))))).....)).))-))..-------.......(((.(((..........................))).))). ( -22.37) >DroSim_CAF1 7617 108 - 1 AAUCGAGUAAUCGGUAUCGAUAA-AAACGCUAGGUGGG---GAGCCUAGCUAAGGAUUUC-GAAU-------UAAGUGUGCCAAGUAACCAAUUAAAAGUAAAUGUAAGAAAACUCGGCG .(((((....))))).((((...-....(((((((...---..)))))))........))-))..-------.......(((.(((..........................))).))). ( -24.03) >DroEre_CAF1 7116 110 - 1 CACAGAGUAAUCGGAAUCGAUAA-AAUGGAUAGGUGGG---AAACCUAGCUAAGGUUUUUUAAAUUGAAAUUUAAUUGCUUAAAAAAGCAGCUUAAAAGUACAUUAAAAA--AUUC---- .........((((....))))..-..(((.(((((...---..))))).))).((((((((((....(..(((((((((((....)))))).)))))..)...)))))))--))).---- ( -18.90) >DroYak_CAF1 8051 86 - 1 CACAGAGUAAUCGGAAUCGAUAAGAAUAGCUAGGUGGG---AAACCUAGCUAAUGUUCUC-AAAU-------UAAAUGUUUCAAAU------------G-----UAGAAG--AUAU---- ....(((..((((....)))).....(((((((((...---..))))))))).....)))-....-------..............------------.-----......--....---- ( -18.70) >consensus CAUAGAGUAAUCGGAAUCGAUAA_AAACGCUAGGUGGG___AAACCUAGCUAAGGUUUUC_GAAU_______UAAAUGUGUCAAGUAACCAAUUAAAAGUAAAUGUAAAAAAACUCG_C_ ..((((((.((((....)))).......(((((((........)))))))..............................................................)))))).. (-13.12 = -13.92 + 0.80)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:06:37 2006