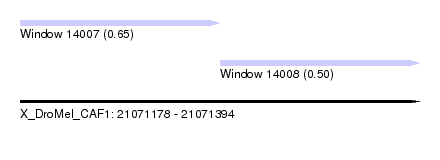
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 21,071,178 – 21,071,394 |
| Length | 216 |
| Max. P | 0.645682 |

| Location | 21,071,178 – 21,071,286 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.12 |
| Mean single sequence MFE | -28.26 |
| Consensus MFE | -25.96 |
| Energy contribution | -26.29 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.645682 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21071178 108 + 22224390 CUCCGCACGACUUCCAAUUUUCAACUCGAAAACUCCAAUUUCGUGUGCAAUUUUCUUGGGUCUGCCG----GAGGAAAAUGCAGAAGGUAGCC--------GGGUGCCAUAUAAAAAAAA ....((((..((..(.((((((.((.(((((.......))))).))(((.(((((((.((....)).----))))))).))).)))))).)..--------))))))............. ( -26.60) >DroEre_CAF1 41613 105 + 1 C--CGCACGACUUCCAAUUUUCAACUCGAAAACUCCAAUUUCGUGUGCAAUUUUCUUGGGUCUGCCG----GCGGAAAAUGCAGAAGGUAGCC--------GGUUGCCAUAUAAAA-ACA .--(((((((.......(((((.....)))))........)))))))...........(((..((((----((.................)))--------))).)))........-... ( -24.59) >DroYak_CAF1 47377 117 + 1 C--CGCACGACUUCCAAUUUUCAACUCGAAAACUCCAAUUUCGUGUGCAAUUUUCUUGGGUCUGCCAAGGGAAGGAAAAUGCAGAAGGUAGCCAUAUAGAAGGCUGCCAUAUAAAA-ACA .--.(((....((((........((.(((((.......))))).))....((..(((((.....)))))..))))))..)))....(((((((........)))))))........-... ( -33.60) >consensus C__CGCACGACUUCCAAUUUUCAACUCGAAAACUCCAAUUUCGUGUGCAAUUUUCUUGGGUCUGCCG____GAGGAAAAUGCAGAAGGUAGCC________GGCUGCCAUAUAAAA_ACA ....((((((.......(((((.....)))))........))))))(((.(((((((.((....)).....))))))).)))....(((((((........)))))))............ (-25.96 = -26.29 + 0.34)

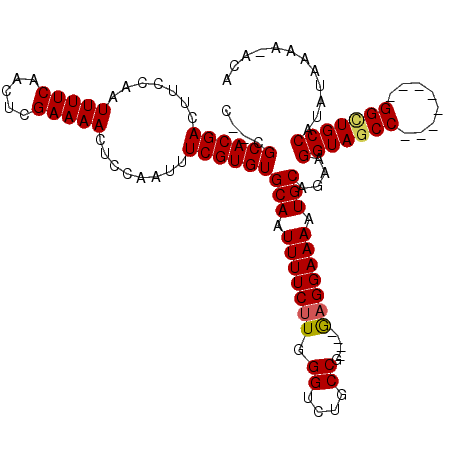

| Location | 21,071,286 – 21,071,394 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.27 |
| Mean single sequence MFE | -17.60 |
| Consensus MFE | -16.67 |
| Energy contribution | -16.57 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.08 |
| Mean z-score | 0.73 |
| Structure conservation index | 0.95 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21071286 108 + 22224390 CAUAGGAAAGGCUAAACGACUUGCCUGAUAACUCGCUAAGCCGCAAAACGUUCCGCAGUUCGUUUCAGUUGUUUUUAUCCCGUACCAUUGUCCUCUGUCCUCGCCAU------------C .(((((((..(((((((((.((((..(((.....((......)).....)))..)))).)))))).)))..))))))).............................------------. ( -17.40) >DroEre_CAF1 41718 119 + 1 CAUAGAAAAGGCUAAACGACUUGCCUGAUAACUCGCUAAGCCGCAAAACGUUCCGCAGUUCGUUUUAGUUGUUUUUAUCCCGUAGCAUU-UCCACUGUCCUCGCCAUGCUUUUCCCCCUU .(((((((..(((((((((.((((..(((.....((......)).....)))..)))).))).))))))..))))))).....(((((.-...............))))).......... ( -19.39) >DroYak_CAF1 47494 99 + 1 CAUAGAAAAGGCUAAACGACUUGCCUGAUAACUCGCUAAGCCGCAAAACGUUCCGCAGUUAGUUUCAGCUGUUUUUAUCCCGCACCAAU-UCCACUGCUU-------------------- .(((((((.((((((((((((.((..(((.....((......)).....)))..)))))).)))).)))).)))))))...(((.....-.....)))..-------------------- ( -16.00) >consensus CAUAGAAAAGGCUAAACGACUUGCCUGAUAACUCGCUAAGCCGCAAAACGUUCCGCAGUUCGUUUCAGUUGUUUUUAUCCCGUACCAUU_UCCACUGUCCUCGCCAU_____________ .(((((((..(((((((((.((((..(((.....((......)).....)))..)))).)))))).)))..))))))).......................................... (-16.67 = -16.57 + -0.11)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:06:09 2006