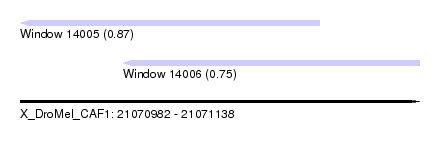
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 21,070,982 – 21,071,138 |
| Length | 156 |
| Max. P | 0.873435 |

| Location | 21,070,982 – 21,071,099 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.16 |
| Mean single sequence MFE | -38.07 |
| Consensus MFE | -29.56 |
| Energy contribution | -30.57 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.59 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.873435 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21070982 117 - 22224390 AACUCCAACCCCUUGCGGCUGCCCCAUUAUCCUGCCACAAUUUUUCCGCACUCCAUAUGUGGAAAAGUGGC---GGGGAAAACAAAUCCAAGCUCUUUAUUCACUCAGCAGUGCAGGCAA ...........((((((.((((.........(((((((...(((((((((.......))))))))))))))---))(((.......)))..................))))))))))... ( -35.80) >DroEre_CAF1 41415 119 - 1 AACUCCGACCCC-UGCGGCUGCCCCAUUAUCCUGCCACAAUUUUUCCACACUCCAUAUGUGGAAAAGUGGCGGCGGGGAAAGCAAAUCCAAGCUCUUCAUUUACUCAGCAGUGCAGGCGA ........(.((-((((.((((........((((((.(.(((((((((((.......))))))))))).).))))))((((((........))).))).........)))))))))).). ( -44.80) >DroYak_CAF1 47180 117 - 1 AACUCCGACCCCUUGCGGCUGCCCCAUUAUCCUACCACAAUAUUUCCACACUCCAUAUGUGGAAAAGUGGC---GGGGGAAGCAAAUCCAAGCUCUUUAUUCACUCAGCAGUGCAAGCAA ...........((((((.((((........((..((((....((((((((.......)))))))).)))).---.))(((......)))..................))))))))))... ( -33.60) >consensus AACUCCGACCCCUUGCGGCUGCCCCAUUAUCCUGCCACAAUUUUUCCACACUCCAUAUGUGGAAAAGUGGC___GGGGAAAGCAAAUCCAAGCUCUUUAUUCACUCAGCAGUGCAGGCAA ...........((((((.(((((((........(((((...(((((((((.......))))))))))))))....)))..(((........))).............))))))))))... (-29.56 = -30.57 + 1.00)



| Location | 21,071,022 – 21,071,138 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.13 |
| Mean single sequence MFE | -45.53 |
| Consensus MFE | -36.46 |
| Energy contribution | -36.80 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.754572 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21071022 116 - 22224390 AGUCCUCGUCGUGG-GCCAGGCCAAAUGCGUCCGGGCUUGAACUCCAACCCCUUGCGGCUGCCCCAUUAUCCUGCCACAAUUUUUCCGCACUCCAUAUGUGGAAAAGUGGC---GGGGAA ..........(.((-((..((((....(((...(((.(((.....))).))).))))))))))))....(((((((((...(((((((((.......))))))))))))))---)))).. ( -44.70) >DroEre_CAF1 41455 118 - 1 AGUCCUCGACCUGG-GCCAGGCCAACUGCGUCCGGGCUUGAACUCCGACCCC-UGCGGCUGCCCCAUUAUCCUGCCACAAUUUUUCCACACUCCAUAUGUGGAAAAGUGGCGGCGGGGAA .(((.(((((((((-(((((.....))).))))))).)))).....)))(((-(((((.....)).......((((((...(((((((((.......))))))))))))))))))))).. ( -49.50) >DroYak_CAF1 47220 117 - 1 AGUCCUCGUCCUGGGGCCAGGCCAAAUGCGUCCAGGCUUGAACUCCGACCCCUUGCGGCUGCCCCAUUAUCCUACCACAAUAUUUCCACACUCCAUAUGUGGAAAAGUGGC---GGGGGA ..(((((((((((((((((((((...........))))))....(((........)))..))))).................((((((((.......)))))))))).)))---))))). ( -42.40) >consensus AGUCCUCGUCCUGG_GCCAGGCCAAAUGCGUCCGGGCUUGAACUCCGACCCCUUGCGGCUGCCCCAUUAUCCUGCCACAAUUUUUCCACACUCCAUAUGUGGAAAAGUGGC___GGGGAA ..(((((....(((.((..((((....(((...(((.(((.....))).))).))))))))).))).......(((((...(((((((((.......))))))))))))))...))))). (-36.46 = -36.80 + 0.34)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:06:07 2006