
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 21,069,708 – 21,070,002 |
| Length | 294 |
| Max. P | 0.995320 |

| Location | 21,069,708 – 21,069,828 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.30 |
| Mean single sequence MFE | -46.92 |
| Consensus MFE | -37.64 |
| Energy contribution | -37.95 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.05 |
| Structure conservation index | 0.80 |
| SVM decision value | 2.27 |
| SVM RNA-class probability | 0.991499 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21069708 120 + 22224390 CCCCGAGAGGGUAUGCAAAACACACAGUCCAAAUCACAACUCACAACUUCUGAGGACGAGAUUUCGUCUGGGCGCUGUCGUGUUGGGCGCUGCUGUCUUAGCGCUUAAGUGCGAUGGCAU .(((....)))...............(((((........((((.......))))((((......)))))))))(((((((((((((((((((......)))))))))).))))))))).. ( -45.60) >DroSim_CAF1 25757 104 + 1 CCCCGAGAGGGUAUGCAAA-------------AUCACAACUCC---UUUCUGAGGACGAGAUUUCGUCUGGGUGCUGUCGUCUUGGGCGCUGCUGUCUUAGCGCUUAAGUGCGAUGGCAU .(((((((((((.((....-------------....)))).))---))))...(((((......))))))))((((((((((((((((((((......))))))))))).))))))))). ( -46.20) >DroEre_CAF1 40187 116 + 1 CCCCGAAAGGGUAUGCUAAGCC-CCAGGCAAAAUCACAACUCC---UUUCCGAGGACGAGAUGUCGUCUGGGUGCUGUCGUCUUGGGCGCUGCUGUCUUAGCGCUUAAGUGCGAUGGCAU .(((....))).(((((.((((-((((((...(((.(...(((---((...))))).).)))...))))))).)))((((((((((((((((......))))))))))).)))))))))) ( -53.10) >DroYak_CAF1 45927 116 + 1 CCACGAAAGGGUAUGUAAGACC-CCAUGCAAAAUCACAACUCC---AUUCUGAGGACGAGAUGUCGUCUGGGUGCUGUCGUCUUGGGCGCUGCUGUCUUAGCGCUUAAGUGCGAUGGCAU ((.(....)))..........(-(((.............(((.---.....)))(((((....)))))))))((((((((((((((((((((......))))))))))).))))))))). ( -42.80) >consensus CCCCGAAAGGGUAUGCAAAACC_CCAGGCAAAAUCACAACUCC___UUUCUGAGGACGAGAUGUCGUCUGGGUGCUGUCGUCUUGGGCGCUGCUGUCUUAGCGCUUAAGUGCGAUGGCAU .(((....)))......................................((..((((((....)))))).)).(((((((((((((((((((......))))))))))).)))))))).. (-37.64 = -37.95 + 0.31)

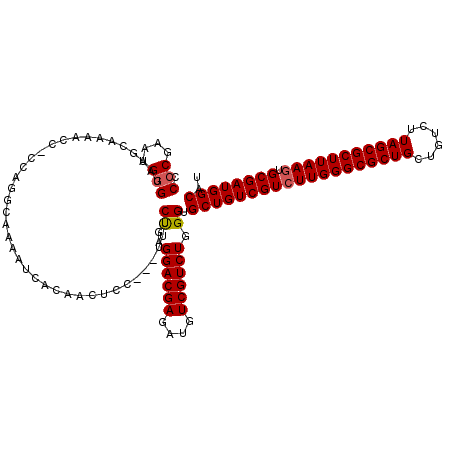
| Location | 21,069,748 – 21,069,868 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.36 |
| Mean single sequence MFE | -43.40 |
| Consensus MFE | -39.31 |
| Energy contribution | -39.38 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.11 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.91 |
| SVM RNA-class probability | 0.982446 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21069748 120 + 22224390 UCACAACUUCUGAGGACGAGAUUUCGUCUGGGCGCUGUCGUGUUGGGCGCUGCUGUCUUAGCGCUUAAGUGCGAUGGCAUUAAGCUAUUAGCAUAAUUUUUGUGCAUUAUCAUUAGGGCC .............(((((......))))).((((((((((((((((((((((......)))))))))).)))))))))............(((((.....)))))............))) ( -39.40) >DroSim_CAF1 25784 117 + 1 UCC---UUUCUGAGGACGAGAUUUCGUCUGGGUGCUGUCGUCUUGGGCGCUGCUGUCUUAGCGCUUAAGUGCGAUGGCAUUAAGCUAUUAGCAUAAUUUUUGUGCAUUAUCAUUAGGGCC .((---((..((((((((......))))).((((((((((((((((((((((......))))))))))).))))))))))).........(((((.....)))))....)))..)))).. ( -45.70) >DroEre_CAF1 40226 117 + 1 UCC---UUUCCGAGGACGAGAUGUCGUCUGGGUGCUGUCGUCUUGGGCGCUGCUGUCUUAGCGCUUAAGUGCGAUGGCAUUAAGCUAUUAGCAUAAUUUUUGUGCAUUAUCAUUAGGGCC .((---((...((((((((....)))))).((((((((((((((((((((((......))))))))))).))))))))))).........(((((.....)))))....))...)))).. ( -43.50) >DroYak_CAF1 45966 117 + 1 UCC---AUUCUGAGGACGAGAUGUCGUCUGGGUGCUGUCGUCUUGGGCGCUGCUGUCUUAGCGCUUAAGUGCGAUGGCAUUAAGCUAUUAGCAUAAUUUUUGUGCAUUAUCAUUUGGGCC (((---(...(((((((((....)))))).((((((((((((((((((((((......))))))))))).))))))))))).........(((((.....)))))....)))..)))).. ( -45.00) >consensus UCC___UUUCUGAGGACGAGAUGUCGUCUGGGUGCUGUCGUCUUGGGCGCUGCUGUCUUAGCGCUUAAGUGCGAUGGCAUUAAGCUAUUAGCAUAAUUUUUGUGCAUUAUCAUUAGGGCC .............((((((....)))))).((((((((((((((((((((((......))))))))))).))))))))............(((((.....)))))............))) (-39.31 = -39.38 + 0.06)



| Location | 21,069,788 – 21,069,887 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.06 |
| Mean single sequence MFE | -34.70 |
| Consensus MFE | -29.40 |
| Energy contribution | -29.65 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.622453 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21069788 99 + 22224390 UGUUGGGCGCUGCUGUCUUAGCGCUUAAGUGCGAUGGCAUUAAGCUAUUAGCAUAAUUUUUGUGCAUUAUCAUUAGGGCCGCGGGGCAAACG-GGGGCGG-------------------- .(((..((.((((.((((((((((....))))((((((.....)))))).(((((.....)))))........)))))).)))).)).))).-.......-------------------- ( -33.80) >DroSim_CAF1 25821 99 + 1 UCUUGGGCGCUGCUGUCUUAGCGCUUAAGUGCGAUGGCAUUAAGCUAUUAGCAUAAUUUUUGUGCAUUAUCAUUAGGGCCGCGGGGCAAAAG-GGGGCGG-------------------- ((((..((.((((.((((((((((....))))((((((.....)))))).(((((.....)))))........)))))).)))).))..)))-)......-------------------- ( -35.00) >DroEre_CAF1 40263 117 + 1 UCUUGGGCGCUGCUGUCUUAGCGCUUAAGUGCGAUGGCAUUAAGCUAUUAGCAUAAUUUUUGUGCAUUAUCAUUAGGGCCGUGGGGCUAA---GGGGCGGUUUGUGUGGGUUUCUGCGGG ((((.(((.(..(.((((((((((....))))((((((.....)))))).(((((.....)))))........)))))).)..).))).)---)))((((.............))))... ( -35.92) >DroYak_CAF1 46003 100 + 1 UCUUGGGCGCUGCUGUCUUAGCGCUUAAGUGCGAUGGCAUUAAGCUAUUAGCAUAAUUUUUGUGCAUUAUCAUUUGGGCCGAGGGGCAAACGCGGGGCGG-------------------- ......((.((((((((((.(.(((((((((.((((((.....)))))).(((((.....))))).....))))))))))..))))))...)))).))..-------------------- ( -34.10) >consensus UCUUGGGCGCUGCUGUCUUAGCGCUUAAGUGCGAUGGCAUUAAGCUAUUAGCAUAAUUUUUGUGCAUUAUCAUUAGGGCCGCGGGGCAAACG_GGGGCGG____________________ .(((((((((((......)))))))))))(((((((((.....)))))).(((((.....)))))............(((....))).........)))..................... (-29.40 = -29.65 + 0.25)



| Location | 21,069,868 – 21,069,966 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.94 |
| Mean single sequence MFE | -33.92 |
| Consensus MFE | -29.41 |
| Energy contribution | -29.60 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.81 |
| Structure conservation index | 0.87 |
| SVM decision value | 2.56 |
| SVM RNA-class probability | 0.995320 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21069868 98 - 22224390 UUGUGUGAGAUGGG-GGCUGAGCCGAAUAUGUCAAUGAUAAAUGGCAUAAUCUGUUUAGCCCACAACAAACUCGCACAAG--------------------CCGCCCC-CGUUUGCCCCGC (((((((((.((.(-((((((((.((.(((((((........))))))).)).)))))))))....))..))))))))).--------------------.......-............ ( -36.90) >DroSim_CAF1 25901 98 - 1 UUGUGUGAGCUGGG-GACUGAGCCGAAUAUGUCAAUGAUAAAUGGCAUAAUCUGUUUAGCCCACAACAAACUCGCACAGG--------------------CCGCCCC-CUUUUGCCCCGC .((((((((.((((-(.((((((.((.(((((((........))))))).)).))))))))).)).....))))))))((--------------------(......-.....))).... ( -33.40) >DroEre_CAF1 40343 116 - 1 UUGUGCGAGAUGGG-GGCUGAGCCGAAUAUGUCAAUGAUAAAUGGCAUAAUCUGUUUAGCCCACAACAAACCCGCACAAACCCGCAGAAACCCACACAAACCGCCCC---UUAGCCCCAC (((((((...((.(-((((((((.((.(((((((........))))))).)).)))))))))....))....))))))).......................((...---...))..... ( -34.20) >DroYak_CAF1 46083 100 - 1 UUGUGUGAGAUGGGCGGCUGAACCGAAUAUGUCAAUGAUAAAUGGCAUAAUCUGUUUAGCCCACAACAAACCCGCACAAA--------------------CCGCCCCGCGUUUGCCCCUC ......(((..((((((((((((.((.(((((((........))))))).)).))))))))...........(((.....--------------------.......)))...))))))) ( -31.20) >consensus UUGUGUGAGAUGGG_GGCUGAGCCGAAUAUGUCAAUGAUAAAUGGCAUAAUCUGUUUAGCCCACAACAAACCCGCACAAA____________________CCGCCCC_CGUUUGCCCCGC (((((((((.((.(.((((((((.((.(((((((........))))))).)).)))))))))....))..)))))))))......................................... (-29.41 = -29.60 + 0.19)

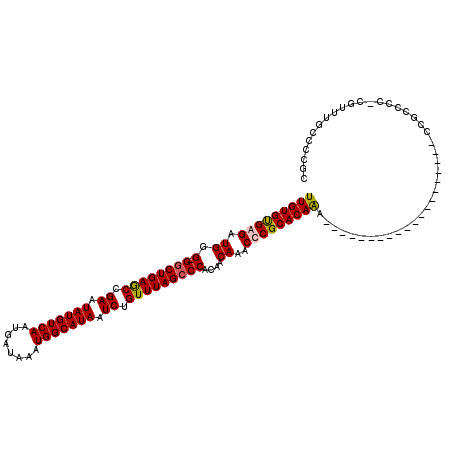

| Location | 21,069,887 – 21,070,002 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.13 |
| Mean single sequence MFE | -38.10 |
| Consensus MFE | -32.46 |
| Energy contribution | -32.15 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.961713 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21069887 115 - 22224390 UCAGCCAUAUUUUCCGGUGUUUGU----AUGUGUUUGCGUUUGUGUGAGAUGGG-GGCUGAGCCGAAUAUGUCAAUGAUAAAUGGCAUAAUCUGUUUAGCCCACAACAAACUCGCACAAG ..((((((((............))----))).))).....(((((((((.((.(-((((((((.((.(((((((........))))))).)).)))))))))....))..))))))))). ( -39.60) >DroSim_CAF1 25920 119 - 1 UCUGCCAUAUUUUCCGGCGUUUGUAUGUAUGUGUUUGCGUUUGUGUGAGCUGGG-GACUGAGCCGAAUAUGUCAAUGAUAAAUGGCAUAAUCUGUUUAGCCCACAACAAACUCGCACAGG ...(((.........)))......(((((......)))))(((((((((.((((-(.((((((.((.(((((((........))))))).)).))))))))).)).....))))))))). ( -37.40) >DroEre_CAF1 40380 115 - 1 UCUGCCAUAUUUUCCGGUGUUUGU----AUGUGUUUGCGUUUGUGCGAGAUGGG-GGCUGAGCCGAAUAUGUCAAUGAUAAAUGGCAUAAUCUGUUUAGCCCACAACAAACCCGCACAAA ..............(((.((((((----.(((.((((((....))))))....(-((((((((.((.(((((((........))))))).)).)))))))))))))))))))))...... ( -39.70) >DroYak_CAF1 46103 120 - 1 UCUGCCAUAUUUUCCGGCGUUUGUAUGUAUGUGUUUGCGUUUGUGUGAGAUGGGCGGCUGAACCGAAUAUGUCAAUGAUAAAUGGCAUAAUCUGUUUAGCCCACAACAAACCCGCACAAA ...(((.........)))...........(((((..(.(((((((((.(.....)((((((((.((.(((((((........))))))).)).))))))))))).))))))).))))).. ( -35.70) >consensus UCUGCCAUAUUUUCCGGCGUUUGU____AUGUGUUUGCGUUUGUGUGAGAUGGG_GGCUGAGCCGAAUAUGUCAAUGAUAAAUGGCAUAAUCUGUUUAGCCCACAACAAACCCGCACAAA ...(((.........))).....................((((((((((.((.(.((((((((.((.(((((((........))))))).)).)))))))))....))..)))))))))) (-32.46 = -32.15 + -0.31)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:06:05 2006