
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 21,068,217 – 21,068,367 |
| Length | 150 |
| Max. P | 0.970546 |

| Location | 21,068,217 – 21,068,327 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.23 |
| Mean single sequence MFE | -33.15 |
| Consensus MFE | -19.67 |
| Energy contribution | -20.80 |
| Covariance contribution | 1.13 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.813710 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
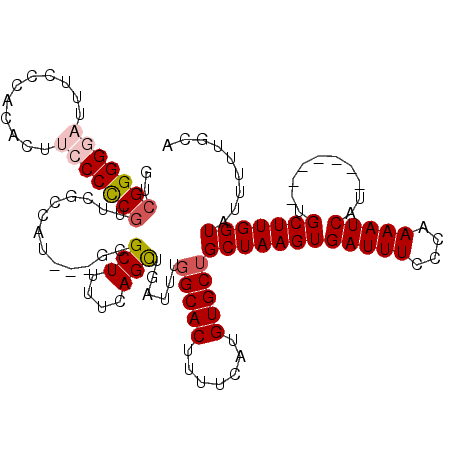
>X_DroMel_CAF1 21068217 110 + 22224390 GUCGGGGGAUUUCCCGCAUUUCCCCCGUUCGCUAU---GCGCUUUUCAGUUGAUUUGGCACUUUUCAUGUGCUGCUAAGUGAUUUCCCAAAAUCAU-------UGCUUGGUAUUUUUACA ..(((((((...........)))))))...((((.---((((...((....))...(((((.......)))))))..((((((((....)))))))-------))).))))......... ( -29.90) >DroSim_CAF1 24344 110 + 1 GUCGGGGGAUUUCCCGCACUUCCCCCGUUCGCUGU---GCGCUUUUCAGCUGAUUUGGCACUUUUCAUGUGCUGCUAAGUGAUUUUCCAAAAUCAU-------UGCUUGGUAUUUUUGCA ..(((((((...........)))))))...((((.---........))))......(((((.......)))))((((((((((((....)))))..-------.)))))))......... ( -31.50) >DroEre_CAF1 38713 116 + 1 GCCGGGGUCC-UUCCACAUUUCCCUCGAUGCCCCU---GCGCUUUUCAGCUGAUUUGGCACUUUUCAUGUGCUGCUAAGUGAUUUCCCAAAAUCAUUUGGAUGUGCUUGGUGUUUUUGCA ((.(((((..-.((............)).))))).---))((......((..(...(((((.......))))).(((((((((((....)))))))))))......)..))......)). ( -31.30) >DroYak_CAF1 44466 120 + 1 GUCGGGGGCUUUUCCACACUUGCCCCCUUCGCCUUUUCGCGCUUUUCAGCUGAUUUGGCACUUUUCAUGUGCAGCUAAGUGAUUUCCCAAAAUCAUUUAGAUGUGCUUGGUAUUUUUUCA ...((((((............))))))...(((...(((.(((....))))))...))).....(((.(..((.(((((((((((....))))))))))).))..).))).......... ( -39.90) >consensus GUCGGGGGAUUUCCCACACUUCCCCCGUUCGCCAU___GCGCUUUUCAGCUGAUUUGGCACUUUUCAUGUGCUGCUAAGUGAUUUCCCAAAAUCAU_______UGCUUGGUAUUUUUGCA ..(((((((...........))))))).............(((....)))......(((((.......)))))((((((((((((....)))))..........)))))))......... (-19.67 = -20.80 + 1.13)

| Location | 21,068,217 – 21,068,327 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.23 |
| Mean single sequence MFE | -29.10 |
| Consensus MFE | -19.17 |
| Energy contribution | -20.67 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.696815 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21068217 110 - 22224390 UGUAAAAAUACCAAGCA-------AUGAUUUUGGGAAAUCACUUAGCAGCACAUGAAAAGUGCCAAAUCAACUGAAAAGCGC---AUAGCGAACGGGGGAAAUGCGGGAAAUCCCCCGAC ..............((.-------.((((((.((.....(((((..((.....))..))))))))))))).((....)).))---........(((((((...........))))))).. ( -26.10) >DroSim_CAF1 24344 110 - 1 UGCAAAAAUACCAAGCA-------AUGAUUUUGGAAAAUCACUUAGCAGCACAUGAAAAGUGCCAAAUCAGCUGAAAAGCGC---ACAGCGAACGGGGGAAGUGCGGGAAAUCCCCCGAC (((..............-------.((((((.((.....(((((..((.....))..)))))))))))))(((....)))))---).......(((((((...........))))))).. ( -30.70) >DroEre_CAF1 38713 116 - 1 UGCAAAAACACCAAGCACAUCCAAAUGAUUUUGGGAAAUCACUUAGCAGCACAUGAAAAGUGCCAAAUCAGCUGAAAAGCGC---AGGGGCAUCGAGGGAAAUGUGGAA-GGACCCCGGC ..........((..((.........((((((.((.....(((((..((.....))..)))))))))))))(((....)))))---.(((((.((.(.(....).).)).-.).)))))). ( -26.90) >DroYak_CAF1 44466 120 - 1 UGAAAAAAUACCAAGCACAUCUAAAUGAUUUUGGGAAAUCACUUAGCUGCACAUGAAAAGUGCCAAAUCAGCUGAAAAGCGCGAAAAGGCGAAGGGGGCAAGUGUGGAAAAGCCCCCGAC ..............(((...((((.((((((....)))))).)))).)))..........((((...((.(((....)))..))...))))..((((((............))))))... ( -32.70) >consensus UGCAAAAAUACCAAGCA_______AUGAUUUUGGGAAAUCACUUAGCAGCACAUGAAAAGUGCCAAAUCAGCUGAAAAGCGC___AAAGCGAACGGGGGAAAUGCGGAAAAGCCCCCGAC ..............((.........((((((.((.....(((((..((.....))..)))))))))))))(((....)))))...........((((((.............)))))).. (-19.17 = -20.67 + 1.50)


| Location | 21,068,254 – 21,068,367 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.20 |
| Mean single sequence MFE | -32.45 |
| Consensus MFE | -25.19 |
| Energy contribution | -25.50 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.10 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.970546 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21068254 113 + 22224390 GCUUUUCAGUUGAUUUGGCACUUUUCAUGUGCUGCUAAGUGAUUUCCCAAAAUCAU-------UGCUUGGUAUUUUUACAACGUUCCCCCGCAUUUUGUGCUAAUAUUGGAGAAGCCUUU (((((((.((((....(((((.......)))))((((((((((((....)))))..-------.))))))).......))))........((((...))))........))))))).... ( -28.90) >DroSim_CAF1 24381 113 + 1 GCUUUUCAGCUGAUUUGGCACUUUUCAUGUGCUGCUAAGUGAUUUUCCAAAAUCAU-------UGCUUGGUAUUUUUGCAACGUUCCCCCGCAUUUUGCGCUAAUAUCAGAGAAGCCUUU ((((((((((......(((((.......)))))....((((((((....)))))))-------))))(((((((..(((((.((........)).)))))..)))))))))))))).... ( -30.90) >DroEre_CAF1 38749 120 + 1 GCUUUUCAGCUGAUUUGGCACUUUUCAUGUGCUGCUAAGUGAUUUCCCAAAAUCAUUUGGAUGUGCUUGGUGUUUUUGCAGCGUUCCCUCGCAUUCUCCGCUAAUAUCCGAGAAGCCUUU (((((((.((((....((((((......(..(..(((((((((((....)))))))))))..)..)..))))))....))))........((.......))........))))))).... ( -35.00) >DroYak_CAF1 44506 120 + 1 GCUUUUCAGCUGAUUUGGCACUUUUCAUGUGCAGCUAAGUGAUUUCCCAAAAUCAUUUAGAUGUGCUUGGUAUUUUUUCAGCACUCCCUCGCAUUUUGCGCUAAUAUCCGAGAAGCCUUU (((((((.(((((...((.(((......(..((.(((((((((((....))))))))))).))..)..))).))...))))).......(((.....))).........))))))).... ( -35.00) >consensus GCUUUUCAGCUGAUUUGGCACUUUUCAUGUGCUGCUAAGUGAUUUCCCAAAAUCAU_______UGCUUGGUAUUUUUGCAACGUUCCCCCGCAUUUUGCGCUAAUAUCCGAGAAGCCUUU (((((((.((((....(((((.......)))))((((((((((((....)))))..........))))))).......)))).......(((.....))).........))))))).... (-25.19 = -25.50 + 0.31)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:05:59 2006