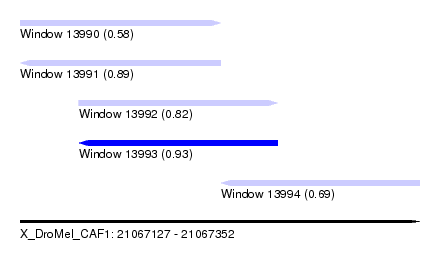
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 21,067,127 – 21,067,352 |
| Length | 225 |
| Max. P | 0.932861 |

| Location | 21,067,127 – 21,067,240 |
|---|---|
| Length | 113 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.92 |
| Mean single sequence MFE | -37.63 |
| Consensus MFE | -23.28 |
| Energy contribution | -24.73 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.580830 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21067127 113 + 22224390 UGGA-------UGUAGGAGCCCUUGUUCCAUUUUUGGCCAUUACCUUUGGCAGUUGCGCCUGGAAAUGACAUUUUCACAUCAUUUGCUGAUGUGCCAAAUGUGUCGUCUGGCUAUCCAGC ((((-------(...(((((....)))))......(((((...((...(((......))).))..(((((((...(((((((.....)))))))......))))))).)))))))))).. ( -37.80) >DroEre_CAF1 37632 115 + 1 UGCGUGUCCUGUGCAGCACCCCUUGGCCCGUUUUUGGCCAUUACCUUUGGCAGUUGCGCCUGGAAAUGACAUUUUCACAUCAUUUGCUGAUGUGGCAAAUG-----UCUGGCUCUCCAGC .((...(((.(((((((...((.(((((.......)))))........))..)))))))..)))...(((((((((((((((.....)))))))).)))))-----))..))........ ( -38.90) >DroYak_CAF1 40995 108 + 1 UGGC-------UGCAGGACCCCUUGUCCCGUUUUUGCCCAUUACCUUUGGCAGUUGCGCCUGGAAAUGACAUUUUCACAUCAUUUGCUGAUGUGGCAAAUG-----UCUGGCUAUCCAGU .(((-------....((((.....)))).((..(((((.(......).)))))..)))))((((...(((((((((((((((.....)))))))).)))))-----))......)))).. ( -36.20) >consensus UGGA_______UGCAGGACCCCUUGUCCCGUUUUUGGCCAUUACCUUUGGCAGUUGCGCCUGGAAAUGACAUUUUCACAUCAUUUGCUGAUGUGGCAAAUG_____UCUGGCUAUCCAGC ((((........(((((.((....)).)).......((((.......))))...)))(((.(((.....(((((((((((((.....)))))))).))))).....))))))..)))).. (-23.28 = -24.73 + 1.45)



| Location | 21,067,127 – 21,067,240 |
|---|---|
| Length | 113 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.92 |
| Mean single sequence MFE | -38.77 |
| Consensus MFE | -24.63 |
| Energy contribution | -24.63 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.49 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.891643 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21067127 113 - 22224390 GCUGGAUAGCCAGACGACACAUUUGGCACAUCAGCAAAUGAUGUGAAAAUGUCAUUUCCAGGCGCAACUGCCAAAGGUAAUGGCCAAAAAUGGAACAAGGGCUCCUACA-------UCCA ...((((((((.......((((((..(((((((.....))))))).))))))...(((((((((....))))...(((....))).....)))))....)))).....)-------))). ( -35.00) >DroEre_CAF1 37632 115 - 1 GCUGGAGAGCCAGA-----CAUUUGCCACAUCAGCAAAUGAUGUGAAAAUGUCAUUUCCAGGCGCAACUGCCAAAGGUAAUGGCCAAAAACGGGCCAAGGGGUGCUGCACAGGACACGCA .(((((((....((-----(((((..(((((((.....))))))).))))))).)))))))((((..(((((...)))..(((((.......)))))))..)))).((.........)). ( -43.10) >DroYak_CAF1 40995 108 - 1 ACUGGAUAGCCAGA-----CAUUUGCCACAUCAGCAAAUGAUGUGAAAAUGUCAUUUCCAGGCGCAACUGCCAAAGGUAAUGGGCAAAAACGGGACAAGGGGUCCUGCA-------GCCA ..((((......((-----(((((..(((((((.....))))))).)))))))...))))(((((...((((...))))....)).....((((((.....))))))..-------))). ( -38.20) >consensus GCUGGAUAGCCAGA_____CAUUUGCCACAUCAGCAAAUGAUGUGAAAAUGUCAUUUCCAGGCGCAACUGCCAAAGGUAAUGGCCAAAAACGGGACAAGGGGUCCUGCA_______CCCA .((((....))))......(((((..(((((((.....))))))).)))))(((((.((.((((....))))...)).)))))..................................... (-24.63 = -24.63 + 0.00)


| Location | 21,067,160 – 21,067,272 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.94 |
| Mean single sequence MFE | -34.43 |
| Consensus MFE | -25.55 |
| Energy contribution | -25.00 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.822266 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21067160 112 + 22224390 UUACCUUUGGCAGUUGCGCCUGGAAAUGACAUUUUCACAUCAUUUGCUGAUGUGCCAAAUGUGUCGUCUGGCUAUCCAGCUAUCCAGCUAUCCAUCU--------CGUUAGCUCUCGAGC .........((....))((.((((.(((((((...(((((((.....)))))))......))))))).(((((....))))))))))).......((--------((........)))). ( -28.70) >DroEre_CAF1 37672 115 + 1 UUACCUUUGGCAGUUGCGCCUGGAAAUGACAUUUUCACAUCAUUUGCUGAUGUGGCAAAUG-----UCUGGCUCUCCAGCUCUUCGGCUCUUCGGCUCUUCAGCUGUUUAGCUCUGGGGC ...((...(((......))).))....(((((((((((((((.....)))))))).)))))-----))(((....)))(((((..((((...(((((....)))))...))))..))))) ( -40.10) >DroYak_CAF1 41028 115 + 1 UUACCUUUGGCAGUUGCGCCUGGAAAUGACAUUUUCACAUCAUUUGCUGAUGUGGCAAAUG-----UCUGGCUAUCCAGUUCUCCAGCUCUUCAGCUCUUCAGCGCUUCAGCUCUCGAGC .....(((((.(((((((((((((...(((((((((((((((.....)))))))).)))))-----))......)))))......((((....)))).....))))...)))).))))). ( -34.50) >consensus UUACCUUUGGCAGUUGCGCCUGGAAAUGACAUUUUCACAUCAUUUGCUGAUGUGGCAAAUG_____UCUGGCUAUCCAGCUCUCCAGCUCUUCAGCUCUUCAGC_CUUUAGCUCUCGAGC ....((..((.(((((.(((.(((.....(((((((((((((.....)))))))).))))).....)))))).....((((....))))...................)))))))..)). (-25.55 = -25.00 + -0.55)


| Location | 21,067,160 – 21,067,272 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.94 |
| Mean single sequence MFE | -36.43 |
| Consensus MFE | -26.29 |
| Energy contribution | -27.07 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.932861 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21067160 112 - 22224390 GCUCGAGAGCUAACG--------AGAUGGAUAGCUGGAUAGCUGGAUAGCCAGACGACACAUUUGGCACAUCAGCAAAUGAUGUGAAAAUGUCAUUUCCAGGCGCAACUGCCAAAGGUAA .((((........))--------)).(((.((((((((..(((((...((((((.......))))))...))))).((((((((....)))))))))))))......))))))....... ( -32.10) >DroEre_CAF1 37672 115 - 1 GCCCCAGAGCUAAACAGCUGAAGAGCCGAAGAGCCGAAGAGCUGGAGAGCCAGA-----CAUUUGCCACAUCAGCAAAUGAUGUGAAAAUGUCAUUUCCAGGCGCAACUGCCAAAGGUAA ((.((.(.(((...(((((...(.((......)))....)))))...)))).((-----(((((..(((((((.....))))))).))))))).......)).))...((((...)))). ( -36.60) >DroYak_CAF1 41028 115 - 1 GCUCGAGAGCUGAAGCGCUGAAGAGCUGAAGAGCUGGAGAACUGGAUAGCCAGA-----CAUUUGCCACAUCAGCAAAUGAUGUGAAAAUGUCAUUUCCAGGCGCAACUGCCAAAGGUAA (((....)))....(((((....((((....))))(((((..(((....)))((-----(((((..(((((((.....))))))).))))))).))))).)))))...((((...)))). ( -40.60) >consensus GCUCGAGAGCUAAAG_GCUGAAGAGCUGAAGAGCUGGAGAGCUGGAUAGCCAGA_____CAUUUGCCACAUCAGCAAAUGAUGUGAAAAUGUCAUUUCCAGGCGCAACUGCCAAAGGUAA (((....)))......................((((((((.((((....))))......(((((..(((((((.....))))))).)))))...))))).((((....))))...))).. (-26.29 = -27.07 + 0.78)


| Location | 21,067,240 – 21,067,352 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.88 |
| Mean single sequence MFE | -33.20 |
| Consensus MFE | -23.73 |
| Energy contribution | -24.30 |
| Covariance contribution | 0.57 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.691289 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21067240 112 - 22224390 UUUUGGGAGUGGCAUUUUGGGCGUUUUUGUAAAUUUCUGGCUACACAGAGAGAAGCGAUACGGGGAGACACUGGCAAAAAGCUCGAGAGCUAACG--------AGAUGGAUAGCUGGAUA (((((....((((.((((((((.(((((((...((((((......)))))).........((((....).)))))))))))))))))))))).))--------))).............. ( -32.50) >DroEre_CAF1 37747 109 - 1 -----------GCGAUUUGGGCGUUUUUGUGAGUUUCUGGCUACACAGAGAGAAGCGAUACAGGGACACACUGGCAAAAGGCCCCAGAGCUAAACAGCUGAAGAGCCGAAGAGCCGAAGA -----------....(((.((((((((((((.((((((..((....))..))))))..)))))))))............(((.(...((((....))))...).))).....))).))). ( -30.80) >DroYak_CAF1 41103 119 - 1 UU-UGGGGGUGGCAUUUUGGGUGUUUUUGUGAAUUUCUGGCUACACAGAGAGAAGCGAUACCGGGAGACACUGGCGAAAAGCUCGAGAGCUGAAGCGCUGAAGAGCUGAAGAGCUGGAGA ((-((..((((...((((.((((((...((...((((((......))))))...)))))))).)))).))))..))))...(((.((..((..(((........)))..))..)).))). ( -36.30) >consensus UU_UGGG_GUGGCAUUUUGGGCGUUUUUGUGAAUUUCUGGCUACACAGAGAGAAGCGAUACAGGGAGACACUGGCAAAAAGCUCGAGAGCUAAAG_GCUGAAGAGCUGAAGAGCUGGAGA .........((((.((((((((.(((((((...((((((......)))))).........((((....).))))))))))))))))))))))............................ (-23.73 = -24.30 + 0.57)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:05:57 2006