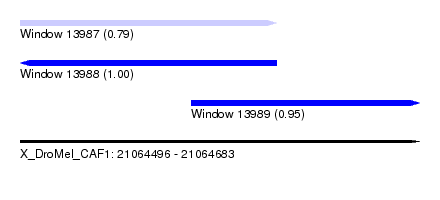
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 21,064,496 – 21,064,683 |
| Length | 187 |
| Max. P | 0.997717 |

| Location | 21,064,496 – 21,064,616 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.84 |
| Mean single sequence MFE | -18.85 |
| Consensus MFE | -9.82 |
| Energy contribution | -10.60 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.787998 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21064496 120 + 22224390 CAAACAUAUAUGCGGGCCAACUUAAAGACUUUCCAGCCAAAUUGCCCGGAAAACAAACUACCAUCUUUACCCCACAAAACACAAAAAAAAAAAAAAAAAAAGGAAGUCAAAUGGUUCCCU .............(((..((((....(((((((..((......))..((...........)).......................................)))))))....))))))). ( -14.10) >DroEre_CAF1 35588 102 + 1 C----AUACAUGCGGGCCAACUUAAAGACUUUCCAGCCAAAUGGCCCGGAAAACAAACUACCAUCUUCCGCCCAAAAAAAAG----------AAGGAAAAAGCAA----ACAUGCUCCCC .----........(((.............(((((.(((....))).(((((..............)))))............----------..))))).((((.----...))))))). ( -18.24) >DroYak_CAF1 38765 112 + 1 C----AUAUAUGGGGGCCAACUUAAAGACUUUCCAGCCAAAUGGCCCGGAAAACAAACUACCAUCUUCCACCCACAAAGAAAAAAAACCAAAAAAAAUAAAGGAA----ACUGGUUCCCU .----......(((((((...........(((((.(((....)))..)))))................................................((...----.))))))))). ( -24.20) >consensus C____AUAUAUGCGGGCCAACUUAAAGACUUUCCAGCCAAAUGGCCCGGAAAACAAACUACCAUCUUCCACCCACAAAAAAAAAAAA__AAAAAAAAAAAAGGAA____ACUGGUUCCCU .............((((((.(.....)..(((((.(((....)))..)))))...........................................................))))))... ( -9.82 = -10.60 + 0.78)

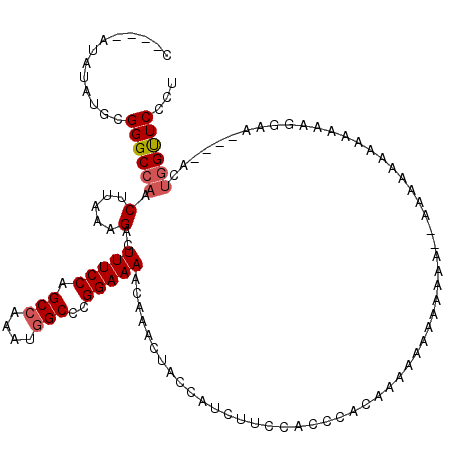
| Location | 21,064,496 – 21,064,616 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.84 |
| Mean single sequence MFE | -32.52 |
| Consensus MFE | -21.03 |
| Energy contribution | -22.03 |
| Covariance contribution | 1.01 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.86 |
| Structure conservation index | 0.65 |
| SVM decision value | 2.92 |
| SVM RNA-class probability | 0.997717 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21064496 120 - 22224390 AGGGAACCAUUUGACUUCCUUUUUUUUUUUUUUUUUUUGUGUUUUGUGGGGUAAAGAUGGUAGUUUGUUUUCCGGGCAAUUUGGCUGGAAAGUCUUUAAGUUGGCCCGCAUAUAUGUUUG ((((((.........)))))).................((((..((((((.(((.......((....((((((((.(.....).)))))))).)).....))).)))))).))))..... ( -25.10) >DroEre_CAF1 35588 102 - 1 GGGGAGCAUGU----UUGCUUUUUCCUU----------CUUUUUUUUGGGCGGAAGAUGGUAGUUUGUUUUCCGGGCCAUUUGGCUGGAAAGUCUUUAAGUUGGCCCGCAUGUAU----G ((..((((...----(((((((((((.(----------((.......))).)))))).)))))..))))..))((((((...((((....)))).......))))))........----. ( -37.30) >DroYak_CAF1 38765 112 - 1 AGGGAACCAGU----UUCCUUUAUUUUUUUUGGUUUUUUUUCUUUGUGGGUGGAAGAUGGUAGUUUGUUUUCCGGGCCAUUUGGCUGGAAAGUCUUUAAGUUGGCCCCCAUAUAU----G ((((((((((.----..............)))))))))).....(((((..((((((..(....)..))))))((((((...((((....)))).......)))))))))))...----. ( -35.16) >consensus AGGGAACCAGU____UUCCUUUUUUUUUUUU__UUUUUCUUUUUUGUGGGCGGAAGAUGGUAGUUUGUUUUCCGGGCCAUUUGGCUGGAAAGUCUUUAAGUUGGCCCGCAUAUAU____G ((((((.........)))))).......................((((...((((((..(....)..))))))((((((...((((....)))).......)))))).))))........ (-21.03 = -22.03 + 1.01)


| Location | 21,064,576 – 21,064,683 |
|---|---|
| Length | 107 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.41 |
| Mean single sequence MFE | -21.33 |
| Consensus MFE | -14.82 |
| Energy contribution | -14.60 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.954832 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21064576 107 + 22224390 ACAAAAAAAAAAAAAAAAAAAGGAAGUCAAAUGGUUCCCUGUUUAUUAAAGCAUUUCUUACGCAUCAUUUGUUGCUUUGCCAGCAAAAAA-GCUGCGUGGAAUGCAAA------------ ................(((.(((.(..(....)..).))).)))......(((((((..(((((...(((.(((((.....))))).)))-..))))))))))))...------------ ( -22.70) >DroEre_CAF1 35664 93 + 1 AG----------AAGGAAAAAGCAA----ACAUGCUCCCCGUUUAUUAAAGCAUUUCUUACGCAUCAUUUGUUGCUUGGCCAACAAAAAA-GCAGAGUGGAAUGC------------CCA ..----------..((....((((.----...))))..............(((((((....((....(((((((......)))))))...-)).....)))))))------------)). ( -17.70) >DroYak_CAF1 38841 116 + 1 AAAAAAACCAAAAAAAAUAAAGGAA----ACUGGUUCCCUGUUUAUUAAAGCAUUUCUUACGCAUCAUUUGUUGCUUUGCCAACAAAAAAGGCAGCGUGGAAUGCAGAUGAAAUGGCCCA ....................((...----.))((..((...((((((...(((((((..((((..((.....))...((((.........))))))))))))))).))))))..)).)). ( -23.60) >consensus AAAAAAA__AAAAAAAAAAAAGGAA____ACUGGUUCCCUGUUUAUUAAAGCAUUUCUUACGCAUCAUUUGUUGCUUUGCCAACAAAAAA_GCAGCGUGGAAUGCA_A_________CCA ..................................................(((((((....((....(((((((......)))))))....)).....)))))))............... (-14.82 = -14.60 + -0.22)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:05:53 2006