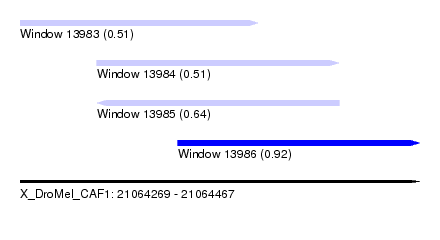
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 21,064,269 – 21,064,467 |
| Length | 198 |
| Max. P | 0.916286 |

| Location | 21,064,269 – 21,064,387 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.62 |
| Mean single sequence MFE | -36.50 |
| Consensus MFE | -30.52 |
| Energy contribution | -31.97 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.84 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.512351 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21064269 118 + 22224390 CCUCGUUUUUU--UUUGCGGCAUGUGGACCGAGCUCCAGCUGGAAGAGGAAGUGCGGCUUGAUUUACUCAACUCGUUUGCUCGCUCGUUUGUUUCCAACUGCUGCAAACAUAUUUUUCUG ...........--(((((((((..((((.((((((((....))).(((.((..(((..((((.....))))..))))).))))))))......))))..)))))))))............ ( -35.60) >DroEre_CAF1 35383 120 + 1 GCUCUCCUUUUUUUUUGGAGCAUGGGGCCCGAGCUCCAGCUGGAAGCGGAAGUGCGGCUUGAUUUACUCAACUCGUUUGCUCGCUCGUUUGUUUCCAACUGCUGCAAACAUAUUUUUGUG .............((((.((((((((((....))))))..((((((((((.(.(((((.(((.((....)).)))...)).))).).))))))))))..)))).))))((((....)))) ( -36.20) >DroYak_CAF1 38534 118 + 1 CCUCUCCUCUU--UUUGCAGCAUGGGACACGAGCUCCAGCUGGAAGCGGAAGUGCGGCUUGAUUUACUCAACUCGUUUGCUCGCUCGUUUGUUUCCAACUGCUGCAAACAUAUUUUUCUG ...........--(((((((((..(((.(((((((((....)))(((((....(((..((((.....))))..)))....))))).)))))).)))...)))))))))............ ( -37.70) >consensus CCUCUCCUUUU__UUUGCAGCAUGGGGCCCGAGCUCCAGCUGGAAGCGGAAGUGCGGCUUGAUUUACUCAACUCGUUUGCUCGCUCGUUUGUUUCCAACUGCUGCAAACAUAUUUUUCUG .............(((((((((((((((....))))))..((((((((((.(.(((((.(((.((....)).)))...)).))).).))))))))))..)))))))))............ (-30.52 = -31.97 + 1.45)



| Location | 21,064,307 – 21,064,427 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.83 |
| Mean single sequence MFE | -30.13 |
| Consensus MFE | -25.25 |
| Energy contribution | -25.70 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.84 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.505632 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21064307 120 + 22224390 UGGAAGAGGAAGUGCGGCUUGAUUUACUCAACUCGUUUGCUCGCUCGUUUGUUUCCAACUGCUGCAAACAUAUUUUUCUGCACCACCCCGGCUAAAAUAUUUCUUAGCCCUUAAACAAAC ((.(((.((..((((((.((((.....))))...((((((..((..(((.......))).)).))))))........))))))..))..((((((........)))))))))...))... ( -27.10) >DroEre_CAF1 35423 119 + 1 UGGAAGCGGAAGUGCGGCUUGAUUUACUCAACUCGUUUGCUCGCUCGUUUGUUUCCAACUGCUGCAAACAUAUUUUUGUGCAGCACCC-GGCUAAAAUAUUUAUUAGCCCUUAAACAAAC ((((((((((.(.(((((.(((.((....)).)))...)).))).).))))))))))..((((((..(((......)))))))))...-((((((........))))))........... ( -35.60) >DroYak_CAF1 38572 120 + 1 UGGAAGCGGAAGUGCGGCUUGAUUUACUCAACUCGUUUGCUCGCUCGUUUGUUUCCAACUGCUGCAAACAUAUUUUUCUGUUCCACCCUGGCUAAAAUAUUUAUUAGCCCUUAAACAAAC (((((.((((((.(((((.(((.((....)).)))...)).))).)((((((...........))))))......))))))))))....((((((........))))))........... ( -27.70) >consensus UGGAAGCGGAAGUGCGGCUUGAUUUACUCAACUCGUUUGCUCGCUCGUUUGUUUCCAACUGCUGCAAACAUAUUUUUCUGCACCACCC_GGCUAAAAUAUUUAUUAGCCCUUAAACAAAC ((((((((((.(.(((((.(((.((....)).)))...)).))).).))))))))))..((.((((............)))).))....((((((........))))))........... (-25.25 = -25.70 + 0.45)



| Location | 21,064,307 – 21,064,427 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.83 |
| Mean single sequence MFE | -35.57 |
| Consensus MFE | -28.73 |
| Energy contribution | -29.73 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.636723 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21064307 120 - 22224390 GUUUGUUUAAGGGCUAAGAAAUAUUUUAGCCGGGGUGGUGCAGAAAAAUAUGUUUGCAGCAGUUGGAAACAAACGAGCGAGCAAACGAGUUGAGUAAAUCAAGCCGCACUUCCUCUUCCA ...........((((((((....))))))))((((.(((((..........((((((.((.((((....).)))..))..))))))(..((((.....))))..)))))).))))..... ( -39.40) >DroEre_CAF1 35423 119 - 1 GUUUGUUUAAGGGCUAAUAAAUAUUUUAGCC-GGGUGCUGCACAAAAAUAUGUUUGCAGCAGUUGGAAACAAACGAGCGAGCAAACGAGUUGAGUAAAUCAAGCCGCACUUCCGCUUCCA ((((((((...((((((........))))))-((.(((((((((......)))..)))))).))..))))))))(((((......((..((((.....))))..))......)))))... ( -35.00) >DroYak_CAF1 38572 120 - 1 GUUUGUUUAAGGGCUAAUAAAUAUUUUAGCCAGGGUGGAACAGAAAAAUAUGUUUGCAGCAGUUGGAAACAAACGAGCGAGCAAACGAGUUGAGUAAAUCAAGCCGCACUUCCGCUUCCA ...........((((((........)))))).((((((((...........((((((.((.((((....).)))..))..))))))(..((((.....))))..)....))))))))... ( -32.30) >consensus GUUUGUUUAAGGGCUAAUAAAUAUUUUAGCC_GGGUGGUGCAGAAAAAUAUGUUUGCAGCAGUUGGAAACAAACGAGCGAGCAAACGAGUUGAGUAAAUCAAGCCGCACUUCCGCUUCCA ...........((((((........))))))..((.(((((..........((((((.((.((((....).)))..))..))))))(..((((.....))))..)))))).))....... (-28.73 = -29.73 + 1.00)


| Location | 21,064,347 – 21,064,467 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.12 |
| Mean single sequence MFE | -28.24 |
| Consensus MFE | -22.89 |
| Energy contribution | -23.00 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.916286 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21064347 120 + 22224390 UCGCUCGUUUGUUUCCAACUGCUGCAAACAUAUUUUUCUGCACCACCCCGGCUAAAAUAUUUCUUAGCCCUUAAACAAACGGAAUAGAUGUGCGGCCCAAUGGAAAACGGAAAACUACAC ....((((((...((((...(((((..((((....(((((.........((((((........))))))..........)))))...)))))))))....)))))))))).......... ( -27.61) >DroEre_CAF1 35463 109 + 1 UCGCUCGUUUGUUUCCAACUGCUGCAAACAUAUUUUUGUGCAGCACCC-GGCUAAAAUAUUUAUUAGCCCUUAAACAAACGGAAUAGAUGUGCGGCCCCAUGCAAA-------AUCG--- .(((.((((((((((....((((((..(((......)))))))))...-((((((........))))))...........)))))))))).)))((.....))...-------....--- ( -31.80) >DroYak_CAF1 38612 113 + 1 UCGCUCGUUUGUUUCCAACUGCUGCAAACAUAUUUUUCUGUUCCACCCUGGCUAAAAUAUUUAUUAGCCCUUAAACAAACGGAAUAGAUGUGCGGCCCCUCGCCAA-------ACUACAC ..((.(((((((...........)))))).......((((((((.....((((((........)))))).((....))..)))))))).).))(((.....)))..-------....... ( -25.30) >consensus UCGCUCGUUUGUUUCCAACUGCUGCAAACAUAUUUUUCUGCACCACCC_GGCUAAAAUAUUUAUUAGCCCUUAAACAAACGGAAUAGAUGUGCGGCCCCAUGCAAA_______ACUACAC ((((.((((((((((....((.((((............)))).))....((((((........))))))...........)))))))))).))))......................... (-22.89 = -23.00 + 0.11)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:05:50 2006