| Sequence ID | X_DroMel_CAF1 |
|---|---|
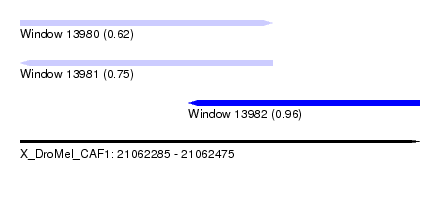
| Location | 21,062,285 – 21,062,475 |
| Length | 190 |
| Max. P | 0.963574 |

| Location | 21,062,285 – 21,062,405 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.13 |
| Mean single sequence MFE | -22.17 |
| Consensus MFE | -19.34 |
| Energy contribution | -18.90 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.622073 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21062285 120 + 22224390 CUUUGCCCCCUAUUUUUUCCUCUAUUUCCUUGACUUUUUCUGUUUUAGUUUUCUUCUAUUUCCCAUUUUGGUGUCAAGUGUUGCGUAUCAUUUAAUGUGACACACUUAGCCGGAAAAUGC ..............((((((.(((.....(((((...........(((.......)))....((.....)).)))))((((..(((........)))..))))...)))..))))))... ( -19.90) >DroEre_CAF1 33412 116 + 1 CUUUGCCCCCCU--UUUUCCUCCAUUUCCUUGGCUU--UCUGUUUUAGUUUUCUUCUUUUGCCCAUUUUGGUGUCAAGUGUUGCGUAUCAUUUAAUGUGACACACUUAGCCGGAAAAUGC ............--..........(((((..((((.--...((...............((((((.....)).).)))((((..(((........)))..))))))..))))))))).... ( -21.40) >DroYak_CAF1 36419 116 + 1 CUUUGCCCCCCA--UUUUCCUCUAUUUCCUUGGCUU--UCAGUUCAAGCUUUCUUCUUUUGCCCAUUUUGGUGUCAAGUGUUGCGUAUCAUUUAAUGUGACACACUUAGCCGGAAAAUGC ..........((--((((((...........(((((--.......)))))...................(((.....((((..(((........)))..)))).....))))))))))). ( -25.20) >consensus CUUUGCCCCCCA__UUUUCCUCUAUUUCCUUGGCUU__UCUGUUUUAGUUUUCUUCUUUUGCCCAUUUUGGUGUCAAGUGUUGCGUAUCAUUUAAUGUGACACACUUAGCCGGAAAAUGC ..............((((((...........((((...........))))...................(((.....((((..(((........)))..)))).....)))))))))... (-19.34 = -18.90 + -0.44)



| Location | 21,062,285 – 21,062,405 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.13 |
| Mean single sequence MFE | -26.53 |
| Consensus MFE | -20.72 |
| Energy contribution | -20.83 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.750921 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21062285 120 - 22224390 GCAUUUUCCGGCUAAGUGUGUCACAUUAAAUGAUACGCAACACUUGACACCAAAAUGGGAAAUAGAAGAAAACUAAAACAGAAAAAGUCAAGGAAAUAGAGGAAAAAAUAGGGGGCAAAG ...((((((..(((.((((((((.......))))))))....((((((.((.....))....(((.......)))...........))))))....))).)))))).............. ( -26.40) >DroEre_CAF1 33412 116 - 1 GCAUUUUCCGGCUAAGUGUGUCACAUUAAAUGAUACGCAACACUUGACACCAAAAUGGGCAAAAGAAGAAAACUAAAACAGA--AAGCCAAGGAAAUGGAGGAAAA--AGGGGGGCAAAG .(((((((.((((..((((((((.......))))))))....(((....((.....))....))).................--.))))..)))))))........--............ ( -24.10) >DroYak_CAF1 36419 116 - 1 GCAUUUUCCGGCUAAGUGUGUCACAUUAAAUGAUACGCAACACUUGACACCAAAAUGGGCAAAAGAAGAAAGCUUGAACUGA--AAGCCAAGGAAAUAGAGGAAAA--UGGGGGGCAAAG .((((((((..(((.((((((((.......))))))))....((((...((.....)).............((((.......--))))))))....))).))))))--)).......... ( -29.10) >consensus GCAUUUUCCGGCUAAGUGUGUCACAUUAAAUGAUACGCAACACUUGACACCAAAAUGGGCAAAAGAAGAAAACUAAAACAGA__AAGCCAAGGAAAUAGAGGAAAA__UGGGGGGCAAAG ....(((((((((..((((((((.......))))))))....(((....((.....))....)))....................))))..)))))........................ (-20.72 = -20.83 + 0.11)



| Location | 21,062,365 – 21,062,475 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.04 |
| Mean single sequence MFE | -32.10 |
| Consensus MFE | -30.67 |
| Energy contribution | -30.23 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.963574 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21062365 110 - 22224390 CGCUUGGCACAGCAAAUUAGCAUUCAGCCAACAUAUGUGCUCAGCUGUCAGUCUGGAAAAGUUCCUCC----------CCGCAUUUUCCGGCUAAGUGUGUCACAUUAAAUGAUACGCAA .(((.((((((........((.....)).......)))))).)))....((.((((((((((......----------..)).))))))))))..((((((((.......)))))))).. ( -31.96) >DroEre_CAF1 33488 118 - 1 CGCUUAGCACAGCAAAUUAGCAUUCAGCCAACA--UGUGCUCAGCUGUCAGUCGGGAAAAGCCCCUCCCCUGUUUUCCCCGCAUUUUCCGGCUAAGUGUGUCACAUUAAAUGAUACGCAA .(((.((((((........((.....)).....--)))))).)))....((((((((((.((..................)).))))))))))..((((((((.......)))))))).. ( -34.69) >DroYak_CAF1 36495 117 - 1 CGCUUAGCACAGCAAAUUAGCAUUCAGCCAACA--UGUGCUCAGCUGUCAGUCUGGAAAAGUCCCUCCCCUGUUUUC-CCGCAUUUUCCGGCUAAGUGUGUCACAUUAAAUGAUACGCAA .(((.((((((........((.....)).....--)))))).)))....((.((((((((((...............-..)).))))))))))..((((((((.......)))))))).. ( -29.65) >consensus CGCUUAGCACAGCAAAUUAGCAUUCAGCCAACA__UGUGCUCAGCUGUCAGUCUGGAAAAGUCCCUCCCCUGUUUUC_CCGCAUUUUCCGGCUAAGUGUGUCACAUUAAAUGAUACGCAA .(((.((((((........((.....)).......)))))).)))....((((.((((((((..................)).))))))))))..((((((((.......)))))))).. (-30.67 = -30.23 + -0.44)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:05:47 2006