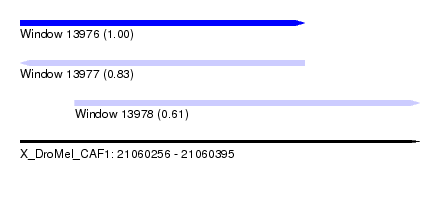
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 21,060,256 – 21,060,395 |
| Length | 139 |
| Max. P | 0.996602 |

| Location | 21,060,256 – 21,060,355 |
|---|---|
| Length | 99 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.20 |
| Mean single sequence MFE | -16.50 |
| Consensus MFE | -16.15 |
| Energy contribution | -15.93 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.98 |
| SVM decision value | 2.72 |
| SVM RNA-class probability | 0.996602 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21060256 99 + 22224390 ---------------------UCCCCCAUCUCCCAAAACCGAUUCCCAUUUCAUAUUGUCCAACAAUUUCAUUUGGGAAUAUGUAUUUCUGGUAUGUCCGAAUAAGCCAAACGCGAAAUG ---------------------.....(((.((.(...((..(((((((......((((.....))))......)))))))..)).....((((.(((....))).))))...).)).))) ( -16.20) >DroEre_CAF1 31372 97 + 1 ---------------------UCCCCCAUCUCCCAAAACCCAUUCCCAUUUCAUAUUGUCCAACAAUUUCAUUUGGGAAUAUGUAUUUCUGGUAUGUCCAAAUAAACCAAC--CGAAAUG ---------------------....................(((((((......((((.....))))......)))))))...((((((((((.(((....))).))))..--.)))))) ( -16.50) >DroYak_CAF1 34321 118 + 1 UCUCCCGUCUCCCAUCUGCCAUCUGCCAUCUCCCAAAACCCAUUCCCAUUUCAUAUUGUCCAACAAUUUCAUUUGGGAAUAUGUAUUUCUGGUAUGUACAAAUAAACCAAC--CGAAAUG .........................................(((((((......((((.....))))......)))))))...((((((((((.(((....))).))))..--.)))))) ( -16.80) >consensus _____________________UCCCCCAUCUCCCAAAACCCAUUCCCAUUUCAUAUUGUCCAACAAUUUCAUUUGGGAAUAUGUAUUUCUGGUAUGUCCAAAUAAACCAAC__CGAAAUG .........................................(((((((......((((.....))))......)))))))...((((((((((.(((....))).)))).....)))))) (-16.15 = -15.93 + -0.22)

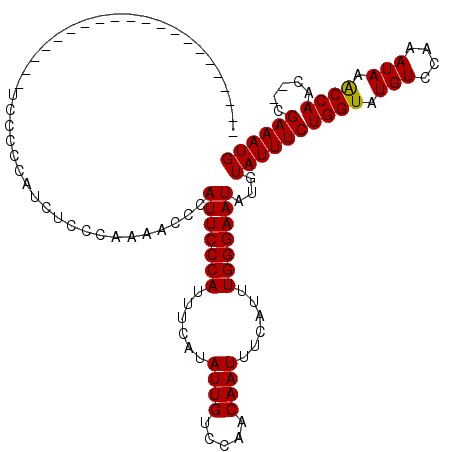
| Location | 21,060,256 – 21,060,355 |
|---|---|
| Length | 99 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.20 |
| Mean single sequence MFE | -26.60 |
| Consensus MFE | -20.92 |
| Energy contribution | -21.70 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.832757 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21060256 99 - 22224390 CAUUUCGCGUUUGGCUUAUUCGGACAUACCAGAAAUACAUAUUCCCAAAUGAAAUUGUUGGACAAUAUGAAAUGGGAAUCGGUUUUGGGAGAUGGGGGA--------------------- ((((((..((((((.....))))))...((((((...(..(((((((..(.(.((((.....)))).).)..))))))).).)))))))))))).....--------------------- ( -24.60) >DroEre_CAF1 31372 97 - 1 CAUUUCG--GUUGGUUUAUUUGGACAUACCAGAAAUACAUAUUCCCAAAUGAAAUUGUUGGACAAUAUGAAAUGGGAAUGGGUUUUGGGAGAUGGGGGA--------------------- ((((((.--(((.(......).)))...((((((...(.((((((((..(.(.((((.....)))).).)..))))))))).)))))))))))).....--------------------- ( -24.70) >DroYak_CAF1 34321 118 - 1 CAUUUCG--GUUGGUUUAUUUGUACAUACCAGAAAUACAUAUUCCCAAAUGAAAUUGUUGGACAAUAUGAAAUGGGAAUGGGUUUUGGGAGAUGGCAGAUGGCAGAUGGGAGACGGGAGA ..((((.--(((..(((((((((.(((.((((((...(.((((((((..(.(.((((.....)))).).)..))))))))).))))))...))).......))))))))).))).)))). ( -30.50) >consensus CAUUUCG__GUUGGUUUAUUUGGACAUACCAGAAAUACAUAUUCCCAAAUGAAAUUGUUGGACAAUAUGAAAUGGGAAUGGGUUUUGGGAGAUGGGGGA_____________________ ((((((...(((.(......).)))...((((((.....((((((((..(.(.((((.....)))).).)..))))))))..)))))))))))).......................... (-20.92 = -21.70 + 0.78)


| Location | 21,060,275 – 21,060,395 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.72 |
| Mean single sequence MFE | -28.83 |
| Consensus MFE | -22.75 |
| Energy contribution | -22.87 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.609508 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21060275 120 + 22224390 GAUUCCCAUUUCAUAUUGUCCAACAAUUUCAUUUGGGAAUAUGUAUUUCUGGUAUGUCCGAAUAAGCCAAACGCGAAAUGUAAAUGUUUCGCGUUCGUUUCGAAGUUGUGCAAGGAAGUA .......(((((...((((...((((((((((((((..((((.........))))..))))))((((..((((((((((......)))))))))).)))).)))))))))))).))))). ( -32.90) >DroEre_CAF1 31391 117 + 1 CAUUCCCAUUUCAUAUUGUCCAACAAUUUCAUUUGGGAAUAUGUAUUUCUGGUAUGUCCAAAUAAACCAAC--CGAAAUGAAAAUGUUUCGCGUUCGCUUCAAACUUGUGCAAAGAAGU- .(((((((......((((.....))))......)))))))....(((((((((.(((....))).)))(((--((((((......)))))).))).((..((....)).))..))))))- ( -23.20) >DroYak_CAF1 34361 117 + 1 CAUUCCCAUUUCAUAUUGUCCAACAAUUUCAUUUGGGAAUAUGUAUUUCUGGUAUGUACAAAUAAACCAAC--CGAAAUGAAAAUGUUUCGCGUUCGUUUCAAAGUUGUGCAAAGAAGU- .(((((((......((((.....))))......)))))))....((((((....(((((((..((((.(((--((((((......)))))).))).)))).....))))))).))))))- ( -30.40) >consensus CAUUCCCAUUUCAUAUUGUCCAACAAUUUCAUUUGGGAAUAUGUAUUUCUGGUAUGUCCAAAUAAACCAAC__CGAAAUGAAAAUGUUUCGCGUUCGUUUCAAAGUUGUGCAAAGAAGU_ .(((((((......((((.....))))......))))))).(((((...((((.(((....))).)))).....((((((..((((.....))))))))))......)))))........ (-22.75 = -22.87 + 0.11)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:05:43 2006