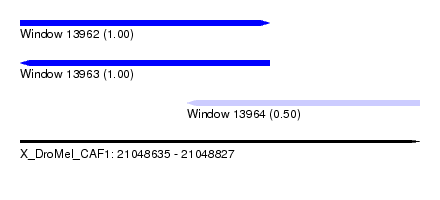
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 21,048,635 – 21,048,827 |
| Length | 192 |
| Max. P | 0.999895 |

| Location | 21,048,635 – 21,048,755 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.32 |
| Mean single sequence MFE | -32.14 |
| Consensus MFE | -31.62 |
| Energy contribution | -31.30 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.98 |
| SVM decision value | 3.32 |
| SVM RNA-class probability | 0.999004 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
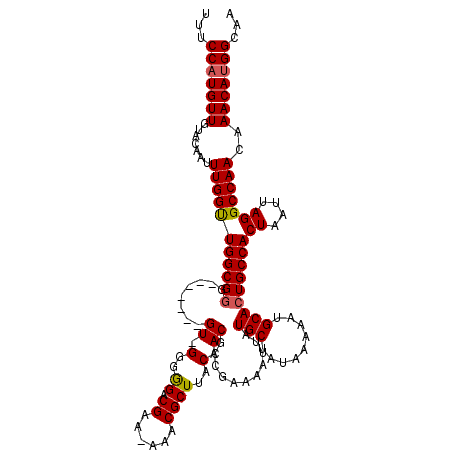
>X_DroMel_CAF1 21048635 120 + 22224390 AAAAAUAGGAUAUAGCCACUAAAAAGUAAUAUGAAAAGGCGGCCAAGAAAACAUUUCUAAGUACGCAUCUGCUGGCGCUUUUGCCAUGUUUGUUGGCCUAAUUAGUGGCAGUGCAUUUUU ..............((((((((...((....(....).))((((((..((((((..........((....)).((((....)))))))))).))))))...))))))))........... ( -32.60) >DroSec_CAF1 19011 118 + 1 --AAAUAGGAUAUGGCCACUAAAAAGUAAUAUCAAAAGGCGGCCAAGGAAACAUUUCUAAGUACGCAUCUGCUGGCGCUUCUGCCAUGUUUGUUGGCCUAAUUAGUGGCAGUGCAUUUUU --.....(.((...((((((((...((...........))((((((.(((.(((..........((....)).((((....)))))))))).))))))...)))))))).)).)...... ( -31.20) >DroSim_CAF1 19578 118 + 1 --AAAUAGGAUAUAGCCACUAAAAAGUAAUAUCAAAAGGCGGCCAAGGAAACAUUUCUAAGUACGCAUCUGCUGGCGCUUCUGCCAUGUUUGUUGGCCUAAUUAGUGGCAGUGCAUUUUU --............((((((((...((...........))((((((.(((.(((..........((....)).((((....)))))))))).))))))...))))))))........... ( -31.10) >DroEre_CAF1 19759 119 + 1 -AAAAUAGGAUAUAGCCACUAAAAAGUAAUACCAAAAGGCGGCCAAGAAAACAUUUCUAAGUACGCAUCUGCCGGCGCUUUUGCCGUGUUUGUUGGCCUAAUUAGUGGCAGUGCAUUUUU -.............((((((((.........((....)).((((((..(((((...........((....))(((((....)))))))))).))))))...))))))))........... ( -34.00) >DroYak_CAF1 21633 120 + 1 AAAAAUAGGAUAUAGCCACUAAAAAGUAAUAUCAAAAGGCGGCCAAGAAAACAUUUCUAAGUACGCAUCUGCUGGCGCUUUUGCCAUGUUUGUUGGCCUAAUUAGUGGCAGUGCAUUUUU ..............((((((((...((...........))((((((..((((((..........((....)).((((....)))))))))).))))))...))))))))........... ( -31.80) >consensus _AAAAUAGGAUAUAGCCACUAAAAAGUAAUAUCAAAAGGCGGCCAAGAAAACAUUUCUAAGUACGCAUCUGCUGGCGCUUUUGCCAUGUUUGUUGGCCUAAUUAGUGGCAGUGCAUUUUU ..............((((((((...((...........))((((((..(((((...........((....))(((((....)))))))))).))))))...))))))))........... (-31.62 = -31.30 + -0.32)



| Location | 21,048,635 – 21,048,755 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.32 |
| Mean single sequence MFE | -31.66 |
| Consensus MFE | -30.04 |
| Energy contribution | -29.72 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.95 |
| SVM decision value | 4.43 |
| SVM RNA-class probability | 0.999895 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21048635 120 - 22224390 AAAAAUGCACUGCCACUAAUUAGGCCAACAAACAUGGCAAAAGCGCCAGCAGAUGCGUACUUAGAAAUGUUUUCUUGGCCGCCUUUUCAUAUUACUUUUUAGUGGCUAUAUCCUAUUUUU ...........((((((((...((((((.(((((((((......))).((....))..........))))))..))))))..................)))))))).............. ( -30.00) >DroSec_CAF1 19011 118 - 1 AAAAAUGCACUGCCACUAAUUAGGCCAACAAACAUGGCAGAAGCGCCAGCAGAUGCGUACUUAGAAAUGUUUCCUUGGCCGCCUUUUGAUAUUACUUUUUAGUGGCCAUAUCCUAUUU-- ...........((((((((..((((((.......))))(((((((((((..((.((((........)))).)).)))).)).))))).......))..))))))))............-- ( -30.40) >DroSim_CAF1 19578 118 - 1 AAAAAUGCACUGCCACUAAUUAGGCCAACAAACAUGGCAGAAGCGCCAGCAGAUGCGUACUUAGAAAUGUUUCCUUGGCCGCCUUUUGAUAUUACUUUUUAGUGGCUAUAUCCUAUUU-- ...........((((((((..((((((.......))))(((((((((((..((.((((........)))).)).)))).)).))))).......))..))))))))............-- ( -30.80) >DroEre_CAF1 19759 119 - 1 AAAAAUGCACUGCCACUAAUUAGGCCAACAAACACGGCAAAAGCGCCGGCAGAUGCGUACUUAGAAAUGUUUUCUUGGCCGCCUUUUGGUAUUACUUUUUAGUGGCUAUAUCCUAUUUU- ...........((((((((...((((((.(((((((((......))))((....))...........)))))..))))))(((....)))........)))))))).............- ( -36.50) >DroYak_CAF1 21633 120 - 1 AAAAAUGCACUGCCACUAAUUAGGCCAACAAACAUGGCAAAAGCGCCAGCAGAUGCGUACUUAGAAAUGUUUUCUUGGCCGCCUUUUGAUAUUACUUUUUAGUGGCUAUAUCCUAUUUUU ...........((((((((...((((((.(((((((((......))).((....))..........))))))..))))))..(....)..........)))))))).............. ( -30.60) >consensus AAAAAUGCACUGCCACUAAUUAGGCCAACAAACAUGGCAAAAGCGCCAGCAGAUGCGUACUUAGAAAUGUUUUCUUGGCCGCCUUUUGAUAUUACUUUUUAGUGGCUAUAUCCUAUUUU_ ...........((((((((...((((((.(((((((((......))))((....))...........)))))..))))))..................)))))))).............. (-30.04 = -29.72 + -0.32)


| Location | 21,048,715 – 21,048,827 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.76 |
| Mean single sequence MFE | -30.92 |
| Consensus MFE | -23.48 |
| Energy contribution | -23.48 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.76 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21048715 112 - 22224390 UUUCCAUGUUGUACAAUUUGGUUGGCGUG-------GU-GGGGGACGAAAAAACGCUUACAACGCCGAAAAUUAUGCUAUAAAAAUGCACUGCCACUAAUUAGGCCAACAAACAUGGCAA ...((((((((((.(((((..(((((((.-------((-(((.(.........).))))).)))))))))))).)))..............(((........))).....)))))))... ( -29.60) >DroSec_CAF1 19089 111 - 1 UUUCCAUGUUGUACAAUUUGGUUGGCGGG-------GU-GGGGGACGAA-AAACGCUUACAACGCCGAAAAUUAUGCUAUAAAAAUGCACUGCCACUAAUUAGGCCAACAAACAUGGCAG ...((((((((((.(((((..((((((..-------((-(((.(.....-...).)))))..))))))))))).)))..............(((........))).....)))))))... ( -30.90) >DroSim_CAF1 19656 111 - 1 UUUCCAUGUUGUACAAUUUGGUUGGCGGG-------GU-GGGGGACGAA-AAACGCUUACAACGCCGAAAAUUAUGCUAUAAAAAUGCACUGCCACUAAUUAGGCCAACAAACAUGGCAG ...((((((((((.(((((..((((((..-------((-(((.(.....-...).)))))..))))))))))).)))..............(((........))).....)))))))... ( -30.90) >DroEre_CAF1 19838 111 - 1 UUUCCAUGUUGUACAAUUUGGCUGGCGGG-------GU-GGGAGACGAA-AAACGCUUACAACGCCGAAAAUUAUGCUAUAAAAAUGCACUGCCACUAAUUAGGCCAACAAACACGGCAA ......((((((.....((((((((((((-------((-(.(((.((..-...)))))....))))........(((.........)))))))))((....))))))).....)))))). ( -29.50) >DroYak_CAF1 21713 119 - 1 UUUCCAUGUUGUACAAUUUGGUUGGCGGGCGAGACGGUGGGGAGACGAA-AAACGCUUACAACGCCGAAAAUUAUGCUAUAAAAAUGCACUGCCACUAAUUAGGCCAACAAACAUGGCAA ...(((((((..........((((((.(((.((.(((((..(((.((..-...)))))....))))).......(((.........)))))))).((....)))))))).)))))))... ( -33.70) >consensus UUUCCAUGUUGUACAAUUUGGUUGGCGGG_______GU_GGGGGACGAA_AAACGCUUACAACGCCGAAAAUUAUGCUAUAAAAAUGCACUGCCACUAAUUAGGCCAACAAACAUGGCAA ...(((((((.......(((((((((((........((.(..((.((......))))..).))...........(((.........)))))))))((....)))))))..)))))))... (-23.48 = -23.48 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:05:31 2006