
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 21,046,386 – 21,046,571 |
| Length | 185 |
| Max. P | 0.975889 |

| Location | 21,046,386 – 21,046,492 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.43 |
| Mean single sequence MFE | -29.30 |
| Consensus MFE | -23.92 |
| Energy contribution | -24.80 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.699886 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21046386 106 - 22224390 GACAUUAUCAACAAGACAAAAAAGCCAGCACAACACCGCUUGGCCCAAAAGUGUUUGUCAUCCAUCCGCCUUUGUCUUCGGUG--------------ACCGGGUGAUAAAUCUGCAUUAA .......................((((((........)).))))......(((((((((((((...((((.........))))--------------...)))))))))))..))..... ( -24.50) >DroSec_CAF1 16771 102 - 1 GACACUAUCAACAAGACGAAAAGGCCAGCACAGCACCGCUUGGCCCAAAAGUGUUUGUCAU----CCGCCUUUGUCUUCGGUG--------------ACCGGGUGAUAAAUCUGCAUUAA ((((((.((........))...(((((((........)).)))))....))))))((((((----(((.....(((......)--------------)))))))))))............ ( -28.20) >DroSim_CAF1 17294 102 - 1 GACAUUAUCAACAAGACAAAAAGGCCAGCACAGCACCGCUUGGCCCAAAAGUGUUUGUCAU----CCGCCUUUGUCUUCGGUG--------------ACCGGGUGAUAAAUCUGCAUUAA ......................(((((((........)).))))).....(((((((((((----(((.....(((......)--------------))))))))))))))..))..... ( -27.20) >DroEre_CAF1 17490 99 - 1 GACAUUAUCAACAAGACAAAGAGGCCAGCUU---ACCGCUUGGCCCAAAAGUGUUUGUCAU----CCGCCUUUGUCUUCGGUG--------------ACCGGGUGAUAAAUCUGCAUUAA ....................(.(((((((..---...)).))))))....(((((((((((----(((.....(((......)--------------))))))))))))))..))..... ( -27.50) >DroYak_CAF1 19282 113 - 1 GGCAUUAUCAACAAGACAAAGAAGCCAGCUC---ACCGCUUGGCCCAAAAGUGUUUGUCAU----CCGCCUUUGUCUUCGGUGACCGGUGACCGGUGACCGGGUGAUAAAUCUGCAUUAA .(((((((((.((((((((((..((((((..---...)).)))).((((....))))....----....)))))))))((((.(((((...))))).))))).))))))...)))..... ( -39.10) >consensus GACAUUAUCAACAAGACAAAAAGGCCAGCACA_CACCGCUUGGCCCAAAAGUGUUUGUCAU____CCGCCUUUGUCUUCGGUG______________ACCGGGUGAUAAAUCUGCAUUAA ((((..........((((....(((((((........)).)))))......))))))))........((.((((((((((((.((((.....)))).)))))).))))))...))..... (-23.92 = -24.80 + 0.88)

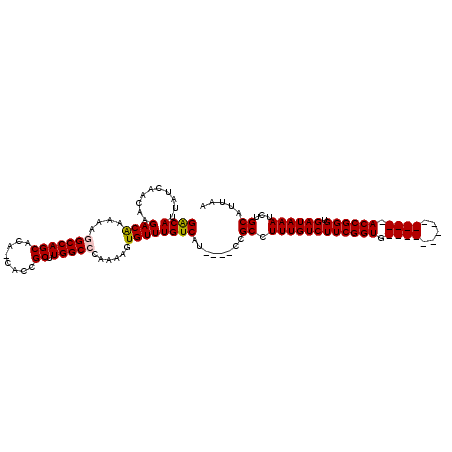
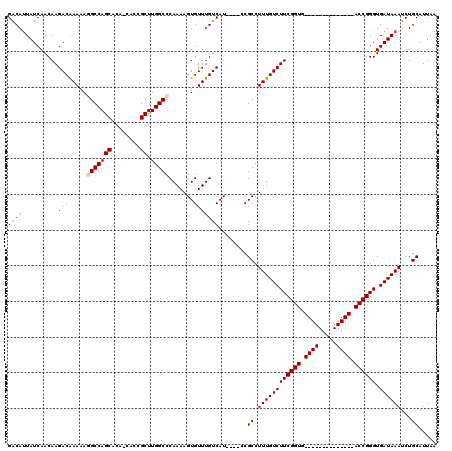
| Location | 21,046,412 – 21,046,531 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.49 |
| Mean single sequence MFE | -33.86 |
| Consensus MFE | -29.26 |
| Energy contribution | -29.86 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.975889 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21046412 119 + 22224390 CGAAGACAAAGGCGGAUGGAUGACAAACACUUUUGGGCCAAGCGGUGUUGUGCUGGCUUUUUUGUCUUGUUGAUAAUGUCAACAAGUUUGAGGAUAAUUCCCUCCACAGA-AAAGUUAAA ......((((((.(..((.....))..).))))))((((((......))).)))((((((((((((((((((((...)))))))))...((((.......)))).)))))-))))))... ( -33.50) >DroSec_CAF1 16797 115 + 1 CGAAGACAAAGGCGG----AUGACAAACACUUUUGGGCCAAGCGGUGCUGUGCUGGCCUUUUCGUCUUGUUGAUAGUGUCAACAAGUUUGGGGAUAAUUCCCUGCACAGA-AAAGUUAAA ((((((((((((.(.----........).))))))(((((.(((......)))))))))))))).(((((((((...)))))))))...(((((....))))).......-......... ( -35.40) >DroSim_CAF1 17320 115 + 1 CGAAGACAAAGGCGG----AUGACAAACACUUUUGGGCCAAGCGGUGCUGUGCUGGCCUUUUUGUCUUGUUGAUAAUGUCAACAAGUUUGGGGAUAAUUCCCUGCACAGA-AAAGUUAAA ......((((((.(.----........).))))))(((((.(((......))))))))((((((((((((((((...)))))))))...(((((....)))))..)))))-))....... ( -35.40) >DroEre_CAF1 17516 112 + 1 CGAAGACAAAGGCGG----AUGACAAACACUUUUGGGCCAAGCGGU---AAGCUGGCCUCUUUGUCUUGUUGAUAAUGUCAACAAGUUUAGGGAUAAUUCCCUGCACAGA-AAAGUAAAA ..((((((((((...----....((((....))))(((((.((...---..)))))))))))))))))((((((...))))))..((.((((((....)))))).))...-......... ( -37.60) >DroYak_CAF1 19322 113 + 1 CGAAGACAAAGGCGG----AUGACAAACACUUUUGGGCCAAGCGGU---GAGCUGGCUUCUUUGUCUUGUUGAUAAUGCCAACCAGUUUAGGGCUAAUUCCCCGCACAAAAAAAGUCAAA ....(((...(((..----....((((....)))).)))..(((((---(((((((.....(((((.....)))))......))))))))(((.....))))))).........)))... ( -27.40) >consensus CGAAGACAAAGGCGG____AUGACAAACACUUUUGGGCCAAGCGGUG_UGUGCUGGCCUUUUUGUCUUGUUGAUAAUGUCAACAAGUUUGGGGAUAAUUCCCUGCACAGA_AAAGUUAAA ..((((((((((...........((((....))))(((((.((........)))))))))))))))))((((((...))))))..((.((((((....)))))).))............. (-29.26 = -29.86 + 0.60)

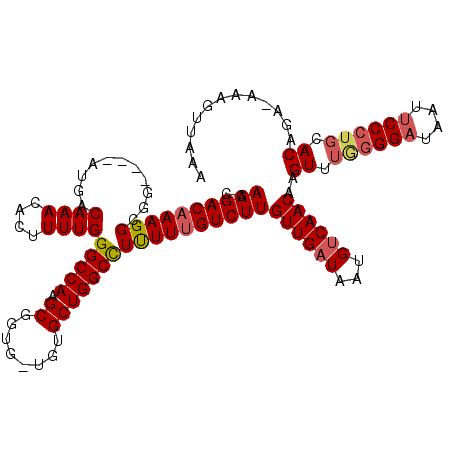
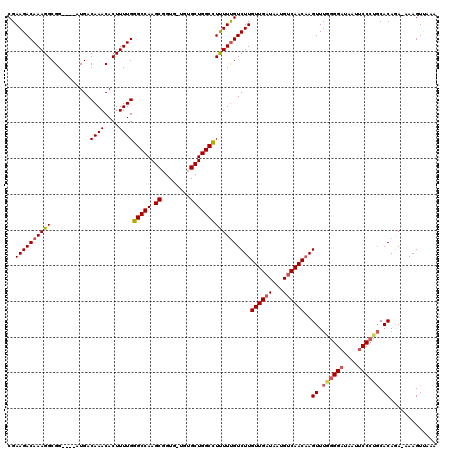
| Location | 21,046,412 – 21,046,531 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.49 |
| Mean single sequence MFE | -28.62 |
| Consensus MFE | -23.64 |
| Energy contribution | -23.56 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.795100 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21046412 119 - 22224390 UUUAACUUU-UCUGUGGAGGGAAUUAUCCUCAAACUUGUUGACAUUAUCAACAAGACAAAAAAGCCAGCACAACACCGCUUGGCCCAAAAGUGUUUGUCAUCCAUCCGCCUUUGUCUUCG ......(((-(.(((.(((((.....)))))...((((((((.....))))))))))).))))((((((........)).)))).((((.(((..((.....))..))).))))...... ( -28.40) >DroSec_CAF1 16797 115 - 1 UUUAACUUU-UCUGUGCAGGGAAUUAUCCCCAAACUUGUUGACACUAUCAACAAGACGAAAAGGCCAGCACAGCACCGCUUGGCCCAAAAGUGUUUGUCAU----CCGCCUUUGUCUUCG .....((((-((......((((....))))....((((((((.....))))))))..))))))((((((........)).)))).((((.(((........----.))).))))...... ( -28.80) >DroSim_CAF1 17320 115 - 1 UUUAACUUU-UCUGUGCAGGGAAUUAUCCCCAAACUUGUUGACAUUAUCAACAAGACAAAAAGGCCAGCACAGCACCGCUUGGCCCAAAAGUGUUUGUCAU----CCGCCUUUGUCUUCG ....(((((-(.(((...((((....))))....((((((((.....)))))))))))....(((((((........)).))))).)))))).........----............... ( -27.70) >DroEre_CAF1 17516 112 - 1 UUUUACUUU-UCUGUGCAGGGAAUUAUCCCUAAACUUGUUGACAUUAUCAACAAGACAAAGAGGCCAGCUU---ACCGCUUGGCCCAAAAGUGUUUGUCAU----CCGCCUUUGUCUUCG .........-...((..(((((....)))))..))(((((((.....)))))))(((((((.(((((((..---...)).)))))((((....))))....----....))))))).... ( -31.90) >DroYak_CAF1 19322 113 - 1 UUUGACUUUUUUUGUGCGGGGAAUUAGCCCUAAACUGGUUGGCAUUAUCAACAAGACAAAGAAGCCAGCUC---ACCGCUUGGCCCAAAAGUGUUUGUCAU----CCGCCUUUGUCUUCG .............((..((((......))))..))..(((((.....)))))(((((((((..((((((..---...)).)))).((((....))))....----....))))))))).. ( -26.30) >consensus UUUAACUUU_UCUGUGCAGGGAAUUAUCCCCAAACUUGUUGACAUUAUCAACAAGACAAAAAGGCCAGCACA_CACCGCUUGGCCCAAAAGUGUUUGUCAU____CCGCCUUUGUCUUCG .............((...(((......)))...))..(((((.....)))))((((((((..(((((((........)).)))))((((....)))).............)))))))).. (-23.64 = -23.56 + -0.08)



| Location | 21,046,452 – 21,046,571 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.35 |
| Mean single sequence MFE | -30.98 |
| Consensus MFE | -14.28 |
| Energy contribution | -15.84 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.46 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.637903 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21046452 119 + 22224390 AGCGGUGUUGUGCUGGCUUUUUUGUCUUGUUGAUAAUGUCAACAAGUUUGAGGAUAAUUCCCUCCACAGA-AAAGUUAAAUCACUACACACGCUUUCGUUCUGUAAGUGAAGUGAAUAAU ((((((((.(((.(((((((((((((((((((((...)))))))))...((((.......)))).)))))-)))))))...))).)))).))))(((((((.......))).)))).... ( -38.40) >DroSec_CAF1 16833 109 + 1 AGCGGUGCUGUGCUGGCCUUUUCGUCUUGUUGAUAGUGUCAACAAGUUUGGGGAUAAUUCCCUGCACAGA-AAAGUUAAAUC-------ACGCUUACAAUCUGUAAAAGAA---CAUAUU ((((...((((((............(((((((((...)))))))))...(((((....))))))))))).-...(......)-------.)))).....(((.....))).---...... ( -29.00) >DroSim_CAF1 17356 109 + 1 AGCGGUGCUGUGCUGGCCUUUUUGUCUUGUUGAUAAUGUCAACAAGUUUGGGGAUAAUUCCCUGCACAGA-AAAGUUAAAUC-------ACGCUUACAAUCUGUAAGUGAA---CAUAUU (((....((((((............(((((((((...)))))))))...(((((....))))))))))).-...))).....-------.(((((((.....)))))))..---...... ( -35.50) >DroEre_CAF1 17552 111 + 1 AGCGGU---AAGCUGGCCUCUUUGUCUUGUUGAUAAUGUCAACAAGUUUAGGGAUAAUUCCCUGCACAGA-AAAGUAAAAUCACUAC--ACGGUUUCAUUCUGCAAAUGAA---AAUAUU .((((.---((((((...(((.((((((((((((...)))))))))...(((((....)))))))).)))-...(((.......)))--.))))))....)))).......---...... ( -28.80) >DroYak_CAF1 19358 112 + 1 AGCGGU---GAGCUGGCUUCUUUGUCUUGUUGAUAAUGCCAACCAGUUUAGGGCUAAUUCCCCGCACAAAAAAAGUCAAAUCACUAG--ACGGUUUCAUUUUGAAAAUGAU---AAUAUU .(((((---(((((((.....(((((.....)))))......))))))))(((.....))))))).........(((.........)--))....(((((.....))))).---...... ( -23.20) >consensus AGCGGUG_UGUGCUGGCCUUUUUGUCUUGUUGAUAAUGUCAACAAGUUUGGGGAUAAUUCCCUGCACAGA_AAAGUUAAAUCACUA___ACGCUUUCAUUCUGUAAAUGAA___AAUAUU ..((((.....))))((........(((((((((...)))))))))...(((((....))))))).((((.(((((...............)))))...))))................. (-14.28 = -15.84 + 1.56)


| Location | 21,046,452 – 21,046,571 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.35 |
| Mean single sequence MFE | -26.92 |
| Consensus MFE | -15.54 |
| Energy contribution | -14.78 |
| Covariance contribution | -0.76 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.668691 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21046452 119 - 22224390 AUUAUUCACUUCACUUACAGAACGAAAGCGUGUGUAGUGAUUUAACUUU-UCUGUGGAGGGAAUUAUCCUCAAACUUGUUGACAUUAUCAACAAGACAAAAAAGCCAGCACAACACCGCU ..........................((((.((((.(((......((((-(.(((.(((((.....)))))...((((((((.....))))))))))).)))))....))).)))))))) ( -31.90) >DroSec_CAF1 16833 109 - 1 AAUAUG---UUCUUUUACAGAUUGUAAGCGU-------GAUUUAACUUU-UCUGUGCAGGGAAUUAUCCCCAAACUUGUUGACACUAUCAACAAGACGAAAAGGCCAGCACAGCACCGCU ......---.....((((.....))))((((-------(..........-.((((((.((((....))))....((((((((.....))))))))............)))))))).))). ( -26.50) >DroSim_CAF1 17356 109 - 1 AAUAUG---UUCACUUACAGAUUGUAAGCGU-------GAUUUAACUUU-UCUGUGCAGGGAAUUAUCCCCAAACUUGUUGACAUUAUCAACAAGACAAAAAGGCCAGCACAGCACCGCU ......---.((((((((.....))))..))-------)).........-.((((((.((((....))))....((((((((.....))))))))............))))))....... ( -27.50) >DroEre_CAF1 17552 111 - 1 AAUAUU---UUCAUUUGCAGAAUGAAACCGU--GUAGUGAUUUUACUUU-UCUGUGCAGGGAAUUAUCCCUAAACUUGUUGACAUUAUCAACAAGACAAAGAGGCCAGCUU---ACCGCU .....(---(((((((...))))))))....--(..((((.((..((((-(.(((..(((((....)))))...((((((((.....))))))))))).)))))..)).))---))..). ( -26.50) >DroYak_CAF1 19358 112 - 1 AAUAUU---AUCAUUUUCAAAAUGAAACCGU--CUAGUGAUUUGACUUUUUUUGUGCGGGGAAUUAGCCCUAAACUGGUUGGCAUUAUCAACAAGACAAAGAAGCCAGCUC---ACCGCU ......---.(((((.....)))))......--...((((.(((.(((((((.((..((((......))))...((.(((((.....))))).))))))))))).))).))---)).... ( -22.20) >consensus AAUAUU___UUCAUUUACAGAAUGAAAGCGU___UAGUGAUUUAACUUU_UCUGUGCAGGGAAUUAUCCCCAAACUUGUUGACAUUAUCAACAAGACAAAAAGGCCAGCACA_CACCGCU .............................................((((...(((...(((......)))....((((((((.....)))))))))))..))))..(((........))) (-15.54 = -14.78 + -0.76)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:05:25 2006