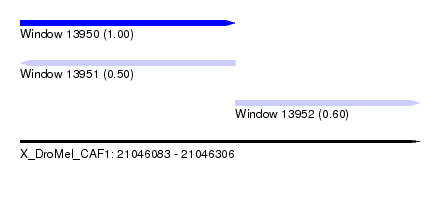
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 21,046,083 – 21,046,306 |
| Length | 223 |
| Max. P | 0.999352 |

| Location | 21,046,083 – 21,046,203 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.33 |
| Mean single sequence MFE | -35.37 |
| Consensus MFE | -30.45 |
| Energy contribution | -31.33 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.84 |
| Structure conservation index | 0.86 |
| SVM decision value | 3.53 |
| SVM RNA-class probability | 0.999352 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21046083 120 + 22224390 ACCCAUUUUUCCGCUUUCCGGCCAAUAUUUAAUUUGUCAAAGUGAGAUUUAACUGUCGGCGGUCCUGCAUCCAGUUGCAGUCGCUAGCACUUAACAAUUUGUUGAUAAAUUAGCCCGAGC ............((((...(((.......(((((((((((...((...((((.(((..((((..(((((......)))))))))..))).))))...))..)))))))))))))).)))) ( -31.81) >DroSec_CAF1 16458 120 + 1 CCCUAUUUUUCCGCUUUCCGGCCAAUAUUUAAUUUGUCAAAGUGAGAUUUCACUGUCGGCGGUCCUGCAUCCAGUUGCAGUCGCUGGCACUUAACAAUUUGUUGAUAAAUUAGCCAGAGU ............(((((..(((.......(((((((((((((((......))))((((((((..(((((......))))))))))))).............))))))))))))))))))) ( -35.61) >DroSim_CAF1 16993 120 + 1 CCCCAUUUAUCCGCUUUCCGGCCAAUAUUUAAUUUGUCAAAGUGAGAUUUCACUGUCGGCGGUCCUGCAUCCAGUUGCAGUCGCUGGCACUUAACAAUUUGUUGAUAAAUUAGCCGGAGU ...............(((((((.......(((((((((((((((......))))((((((((..(((((......))))))))))))).............)))))))))))))))))). ( -41.61) >DroEre_CAF1 17214 120 + 1 CCCCAUUUUUCCGCCUUCCGGCUAAUAUUUAAUUUGUCAAAGUGAGAUUUCACUGUCGGCGGUCCUGCUUCCAGUUGCAGUCGCUUGCACUUAACAAUUUGUUGAUAAAUUAGCCGGAGU ...............(((((((((((.......(((((((((((......))))((.(((((..((((........))))))))).)).............)))))))))))))))))). ( -36.21) >DroYak_CAF1 19000 120 + 1 ACCCAUUUUUCCGCUUUCCGGCUAAUAUUUAAUUUGUCAAAGUGAGAUUUCACUGUCGGCGGUCCUGCCUCCAGUUGCAGUCGCCUGCACUUAACAAUUUGUUGAUAAAUUAGCUGAAGC ............((((..((((((((..(((((((((..(((((.((((.(((((..((((....))))..))).)).)))).....))))).))))...)))))...)))))))))))) ( -31.60) >consensus CCCCAUUUUUCCGCUUUCCGGCCAAUAUUUAAUUUGUCAAAGUGAGAUUUCACUGUCGGCGGUCCUGCAUCCAGUUGCAGUCGCUGGCACUUAACAAUUUGUUGAUAAAUUAGCCGGAGU ...............(((((((.......(((((((((((((((......))))((.(((((..(((((......)))))))))).)).............)))))))))))))))))). (-30.45 = -31.33 + 0.88)

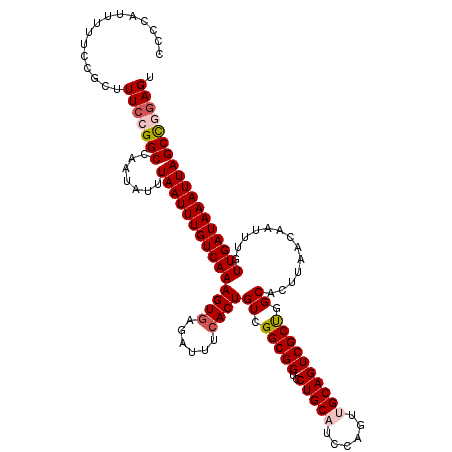

| Location | 21,046,083 – 21,046,203 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.33 |
| Mean single sequence MFE | -30.75 |
| Consensus MFE | -27.03 |
| Energy contribution | -27.11 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.15 |
| Structure conservation index | 0.88 |
| SVM decision value | -0.08 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21046083 120 - 22224390 GCUCGGGCUAAUUUAUCAACAAAUUGUUAAGUGCUAGCGACUGCAACUGGAUGCAGGACCGCCGACAGUUAAAUCUCACUUUGACAAAUUAAAUAUUGGCCGGAAAGCGGAAAAAUGGGU ((((.(((((((...........((((((((((...(((.(((((......)))))...))).((........)).))).))))))).......))))))).))..))............ ( -26.67) >DroSec_CAF1 16458 120 - 1 ACUCUGGCUAAUUUAUCAACAAAUUGUUAAGUGCCAGCGACUGCAACUGGAUGCAGGACCGCCGACAGUGAAAUCUCACUUUGACAAAUUAAAUAUUGGCCGGAAAGCGGAAAAAUAGGG ..((((((((((...........((((((((((.(.(((.(((((......)))))...))).).))((((....)))))))))))).......))))))))))................ ( -30.47) >DroSim_CAF1 16993 120 - 1 ACUCCGGCUAAUUUAUCAACAAAUUGUUAAGUGCCAGCGACUGCAACUGGAUGCAGGACCGCCGACAGUGAAAUCUCACUUUGACAAAUUAAAUAUUGGCCGGAAAGCGGAUAAAUGGGG ..((((((((((...........((((((((((.(.(((.(((((......)))))...))).).))((((....)))))))))))).......))))))))))................ ( -32.67) >DroEre_CAF1 17214 120 - 1 ACUCCGGCUAAUUUAUCAACAAAUUGUUAAGUGCAAGCGACUGCAACUGGAAGCAGGACCGCCGACAGUGAAAUCUCACUUUGACAAAUUAAAUAUUAGCCGGAAGGCGGAAAAAUGGGG ..((((((((((...........(((((((......(((.((((........))))...)))....(((((....)))))))))))).......))))))))))................ ( -31.37) >DroYak_CAF1 19000 120 - 1 GCUUCAGCUAAUUUAUCAACAAAUUGUUAAGUGCAGGCGACUGCAACUGGAGGCAGGACCGCCGACAGUGAAAUCUCACUUUGACAAAUUAAAUAUUAGCCGGAAAGCGGAAAAAUGGGU (((((.((((((...........(((((((((((((....)))))((((..(((......)))..))))..........)))))))).......))))))..).))))............ ( -32.57) >consensus ACUCCGGCUAAUUUAUCAACAAAUUGUUAAGUGCAAGCGACUGCAACUGGAUGCAGGACCGCCGACAGUGAAAUCUCACUUUGACAAAUUAAAUAUUGGCCGGAAAGCGGAAAAAUGGGG ..((((((((((...........(((((((......(((.((((........))))...)))....(((((....)))))))))))).......))))))))))................ (-27.03 = -27.11 + 0.08)


| Location | 21,046,203 – 21,046,306 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 77.98 |
| Mean single sequence MFE | -37.45 |
| Consensus MFE | -23.39 |
| Energy contribution | -24.95 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.603321 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21046203 103 + 22224390 UGGAGC------UGGAGCUGGAGCCGGAUCCAGAGUC------AGAGUCAACGCCGAAGUCAAUCUCACCGACGAAGGGA---GAGUGGCUUAAAAAGUUGCUUUCCUCUUGGCCGGC (((..(------(((..(((((......)))))..))------))..)))..((((..(((.........))).((((((---(((..(((.....)))..)))))).)))...)))) ( -40.00) >DroSec_CAF1 16578 115 + 1 CGGAGCCGAAGCCGGAGCCGGAGGCGGAUCCAGAGUCAGAGUCAGAGUCAACGCCAAAGUCAAUCUCGCUGACGAAGGGA---GAGUGGCUUAAAAAGUUGCUUUCCUUUUGGCCGGC ....((((..((((((.(((....))).)))...(((((.....(((...((......))....))).)))))(((((((---(((..(((.....)))..))))))))))))))))) ( -46.00) >DroSim_CAF1 17113 103 + 1 CGGAGG------CGGAGCCGGAGGCGGAUCCAGAGUC------AGAGUCAACGCCAAAGUCAAUCUCACUGACGAAGGGA---GAGUGGCUUAAAAAGUUGCUUUCCUUUUGGCCGGC ....((------((((.(((....))).)))..(((.------(((....((......))...))).)))..((((((((---(((..(((.....)))..))))))))))))))... ( -38.10) >DroYak_CAF1 19120 84 + 1 CGG----------------------GGAUCCA------------AAGCCAACGCCAAAGUCAAUCUCACUGACGGCGGGAGUAGAGUGGCUUAAAAAGUUGCUUUCCUUUUGGCCGGC .((----------------------....)).------------..(((((((((...((((.......)))))))).(((.((((..(((.....)))..)))).)))))))).... ( -25.70) >consensus CGGAGC______CGGAGCCGGAGGCGGAUCCAGAGUC______AGAGUCAACGCCAAAGUCAAUCUCACUGACGAAGGGA___GAGUGGCUUAAAAAGUUGCUUUCCUUUUGGCCGGC .............(((.(((....))).)))...............(((...(((...((((.......))))(((((((...(((..(((.....)))..))))))))))))).))) (-23.39 = -24.95 + 1.56)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:05:20 2006