
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 21,045,666 – 21,046,003 |
| Length | 337 |
| Max. P | 0.991764 |

| Location | 21,045,666 – 21,045,786 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.83 |
| Mean single sequence MFE | -30.24 |
| Consensus MFE | -26.80 |
| Energy contribution | -26.92 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.71 |
| SVM RNA-class probability | 0.973556 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21045666 120 + 22224390 UUAAAUUUAUGACGAAACGCAAAUCGCAUAAAAAGGUUUGCUCCGUCUCGCUCACACUAUGUGUUGCCAUUUCACUUGCGCCCACUGUUUUGUGGUCGCAUAAAAAAUGCGGCAAAUAUU .........((((((.(((((((((.........)))))))...)).))).)))...(((.((((((.((((....((((.((((......)))).))))....)))))))))).))).. ( -32.00) >DroSec_CAF1 16058 120 + 1 UUAAAUUUAUGACGAAACGCAAAUCGCAUAAAAAGGCGUGCUCUGUCUUGCUCGCACUAUGUGUUGCCAUUUCACUUGCGCCCACUGUUUUGUGGUCGCAUAAAAAAUGCGGCAAAUAUU .....((((((.(((........))))))))).(((((.((........)).))).))...((((((.((((....((((.((((......)))).))))....))))))))))...... ( -34.30) >DroSim_CAF1 16589 120 + 1 UUAAAUUUAUGACGAAACGCAAAUCGCAUAAAAAGGCUUGCUCUGUCUCGCUCGCACUAUGUGUUGCCAUUUCACUUGCGCCCACUGUUUUGUGGUCGCAUAAAAAAUGCGGCAAAUAUU .....((((((.(((........))))))))).((((..((........))..)).))...((((((.((((....((((.((((......)))).))))....))))))))))...... ( -30.50) >DroEre_CAF1 16857 120 + 1 UUAAAUUUAUGACGAAACGCAAAUCGCAUAAAAAGGUCUGCCCCGUCCCGCUCGCACUGUGUGCUGCUAUUUCACUUGCACCCACUGUUUUGUGGUCGCAUAAAAAAUGCGGCAAAUAUU ..........((((....(((.(((.........))).)))..)))).(((.......)))((((((.((((....(((..((((......))))..)))....))))))))))...... ( -28.50) >DroYak_CAF1 18589 120 + 1 UUAAAUUUAUGACGAAACGCAAAUCGCAUAAAAAGGUAUGCUCCGUCUCGCUCACACGGUGUGUUGCUAUUUCACUCCCUCCCACUGUUUUGUGGUCGCAUAAAAAAUGCGGCAAAUAUU .....((((((.(((........)))))))))..(((((((.((((.........)))).))).))))..............(((......)))(((((((.....)))))))....... ( -25.90) >consensus UUAAAUUUAUGACGAAACGCAAAUCGCAUAAAAAGGUUUGCUCCGUCUCGCUCGCACUAUGUGUUGCCAUUUCACUUGCGCCCACUGUUUUGUGGUCGCAUAAAAAAUGCGGCAAAUAUU .....((((((.(((........)))))))))...((..((........))..))..(((.((((((.((((....((((.((((......)))).))))....)))))))))).))).. (-26.80 = -26.92 + 0.12)



| Location | 21,045,666 – 21,045,786 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.83 |
| Mean single sequence MFE | -32.62 |
| Consensus MFE | -28.28 |
| Energy contribution | -28.04 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.897375 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21045666 120 - 22224390 AAUAUUUGCCGCAUUUUUUAUGCGACCACAAAACAGUGGGCGCAAGUGAAAUGGCAACACAUAGUGUGAGCGAGACGGAGCAAACCUUUUUAUGCGAUUUGCGUUUCGUCAUAAAUUUAA .....(((((..(((((...((((.((((......)))).))))...)))))))))).......(((((.(((((((.((((..........)))....).))))))))))))....... ( -32.60) >DroSec_CAF1 16058 120 - 1 AAUAUUUGCCGCAUUUUUUAUGCGACCACAAAACAGUGGGCGCAAGUGAAAUGGCAACACAUAGUGCGAGCAAGACAGAGCACGCCUUUUUAUGCGAUUUGCGUUUCGUCAUAAAUUUAA .....((((((((((.....((((.((((......)))).)))).......((....))...)))))).))))((((((((((((........)))...))).))).))).......... ( -32.80) >DroSim_CAF1 16589 120 - 1 AAUAUUUGCCGCAUUUUUUAUGCGACCACAAAACAGUGGGCGCAAGUGAAAUGGCAACACAUAGUGCGAGCGAGACAGAGCAAGCCUUUUUAUGCGAUUUGCGUUUCGUCAUAAAUUUAA ....(((((((((((.....((((.((((......)))).)))).......((....))...)))))).)))))..............(((((((((........))).))))))..... ( -31.90) >DroEre_CAF1 16857 120 - 1 AAUAUUUGCCGCAUUUUUUAUGCGACCACAAAACAGUGGGUGCAAGUGAAAUAGCAGCACACAGUGCGAGCGGGACGGGGCAGACCUUUUUAUGCGAUUUGCGUUUCGUCAUAAAUUUAA ....((((((.((((((...((((.((((......)))).))))...))))).((.((((...))))..)).....).))))))....(((((((((........))).))))))..... ( -34.40) >DroYak_CAF1 18589 120 - 1 AAUAUUUGCCGCAUUUUUUAUGCGACCACAAAACAGUGGGAGGGAGUGAAAUAGCAACACACCGUGUGAGCGAGACGGAGCAUACCUUUUUAUGCGAUUUGCGUUUCGUCAUAAAUUUAA .........(((((.....))))).((((......))))...((.(((.........))).)).(((((.(((((((.((((((......)))))....).))))))))))))....... ( -31.40) >consensus AAUAUUUGCCGCAUUUUUUAUGCGACCACAAAACAGUGGGCGCAAGUGAAAUGGCAACACAUAGUGCGAGCGAGACGGAGCAAACCUUUUUAUGCGAUUUGCGUUUCGUCAUAAAUUUAA ....(((((((((((.....((((.((((......)))).)))).(((.........)))..)))))).)))))..............(((((((((........))).))))))..... (-28.28 = -28.04 + -0.24)


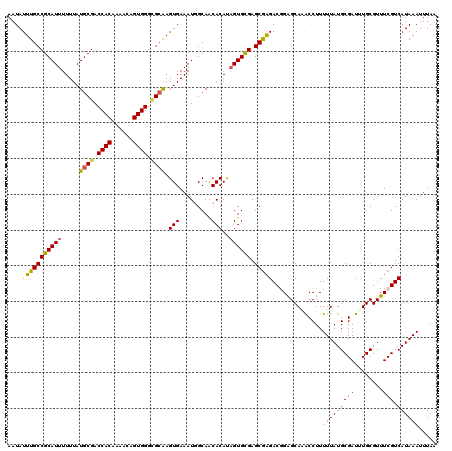
| Location | 21,045,706 – 21,045,826 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.50 |
| Mean single sequence MFE | -32.16 |
| Consensus MFE | -26.64 |
| Energy contribution | -26.88 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.704071 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21045706 120 + 22224390 CUCCGUCUCGCUCACACUAUGUGUUGCCAUUUCACUUGCGCCCACUGUUUUGUGGUCGCAUAAAAAAUGCGGCAAAUAUUUGCGUGCAGCUGCGACCCUUCAAACCCAACGGAAAACCGA ....(((.(((((((.((((.((((((.((((....((((.((((......)))).))))....)))))))))).)))...).))).))).).))).............(((....))). ( -32.70) >DroSec_CAF1 16098 118 + 1 CUCUGUCUUGCUCGCACUAUGUGUUGCCAUUUCACUUGCGCCCACUGUUUUGUGGUCGCAUAAAAAAUGCGGCAAAUAUUUGAGUGCAGCUGCGACCCUUCAAACCCCACGGAAAACG-- ....(((..(((.(((((..((((((((((((....((((.((((......)))).))))....))))..))).)))))...))))))))...)))........((....))......-- ( -33.70) >DroSim_CAF1 16629 118 + 1 CUCUGUCUCGCUCGCACUAUGUGUUGCCAUUUCACUUGCGCCCACUGUUUUGUGGUCGCAUAAAAAAUGCGGCAAAUAUUUGAGUGCAGCUGCGACCCUUCAAACCCCACGGAAAACG-- .(((((...(((.(((((..((((((((((((....((((.((((......)))).))))....))))..))).)))))...))))))))...((....)).......))))).....-- ( -34.00) >DroEre_CAF1 16897 116 + 1 CCCCGUCCCGCUCGCACUGUGUGCUGCUAUUUCACUUGCACCCACUGUUUUGUGGUCGCAUAAAAAAUGCGGCAAAUAUUUGCGUGCAGCUGCGACCCUUCAAACCCAUCGGAAAA---- ..(((....((.((((.(((.((((((.((((....(((..((((......))))..)))....)))))))))).)))..)))).))......((....))........)))....---- ( -32.60) >DroYak_CAF1 18629 118 + 1 CUCCGUCUCGCUCACACGGUGUGUUGCUAUUUCACUCCCUCCCACUGUUUUGUGGUCGCAUAAAAAAUGCGGCAAAUAUUUGCGUGCAGCUGCGACCCUUCAAACCCAUCGGAAAAUA-- .((((....((.(((.....)))..)).....................((((.(((((((.......((((((((....)))).))))..)))))))...)))).....)))).....-- ( -27.80) >consensus CUCCGUCUCGCUCGCACUAUGUGUUGCCAUUUCACUUGCGCCCACUGUUUUGUGGUCGCAUAAAAAAUGCGGCAAAUAUUUGCGUGCAGCUGCGACCCUUCAAACCCAACGGAAAACG__ ....(((.(((((((.(....((((((.((((....((((.((((......)))).))))....)))))))))).......).))).))).).)))........((....))........ (-26.64 = -26.88 + 0.24)



| Location | 21,045,746 – 21,045,866 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.46 |
| Mean single sequence MFE | -33.18 |
| Consensus MFE | -28.28 |
| Energy contribution | -29.08 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.903766 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21045746 120 + 22224390 CCCACUGUUUUGUGGUCGCAUAAAAAAUGCGGCAAAUAUUUGCGUGCAGCUGCGACCCUUCAAACCCAACGGAAAACCGAACGAAACUGAACGGAACGGAAAAUGUUGACAGUGGGUAGC ((((((((((((.(((((((.......((((((((....)))).))))..)))))))...))))..(((((.....(((..((........))...)))....))))))))))))).... ( -40.20) >DroSec_CAF1 16138 110 + 1 CCCACUGUUUUGUGGUCGCAUAAAAAAUGCGGCAAAUAUUUGAGUGCAGCUGCGACCCUUCAAACCCCACGGAAAACG----------GAACGGAACGGAAAAUGUUGACAGCGGGUAGC (((.((((((((((((((((.......((((.(((....)))..))))..))))))).......((....))...)))----------))))))((((.....))))......))).... ( -29.10) >DroSim_CAF1 16669 110 + 1 CCCACUGUUUUGUGGUCGCAUAAAAAAUGCGGCAAAUAUUUGAGUGCAGCUGCGACCCUUCAAACCCCACGGAAAACG----------GAACGGAACGGAAAAUGUUGACAGCGGGUAGC (((.((((((((((((((((.......((((.(((....)))..))))..))))))).......((....))...)))----------))))))((((.....))))......))).... ( -29.10) >DroEre_CAF1 16937 93 + 1 CCCACUGUUUUGUGGUCGCAUAAAAAAUGCGGCAAAUAUUUGCGUGCAGCUGCGACCCUUCAAACCCAUCGGAAAA---------------------------UGUUGACAGUGGGUAGC ((((((((((((.(((((((.......((((((((....)))).))))..)))))))...))).((....))....---------------------------....))))))))).... ( -33.00) >DroYak_CAF1 18669 110 + 1 CCCACUGUUUUGUGGUCGCAUAAAAAAUGCGGCAAAUAUUUGCGUGCAGCUGCGACCCUUCAAACCCAUCGGAAAAUA----------GAACGGAACGGAAAUUGUUGACAGUGGGUAGC ((((((((((((.(((((((.......((((((((....)))).))))..)))))))...))).((..(((.......----------...)))...))........))))))))).... ( -34.50) >consensus CCCACUGUUUUGUGGUCGCAUAAAAAAUGCGGCAAAUAUUUGCGUGCAGCUGCGACCCUUCAAACCCAACGGAAAACG__________GAACGGAACGGAAAAUGUUGACAGUGGGUAGC ((((((((((((.(((((((.......((((((((....)))).))))..)))))))...))).((....))...................................))))))))).... (-28.28 = -29.08 + 0.80)


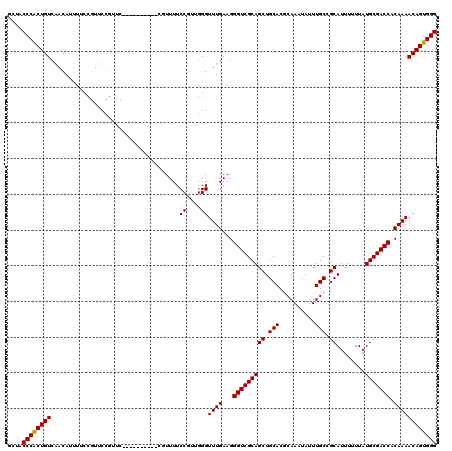
| Location | 21,045,746 – 21,045,866 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.46 |
| Mean single sequence MFE | -33.04 |
| Consensus MFE | -28.78 |
| Energy contribution | -28.54 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.868183 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21045746 120 - 22224390 GCUACCCACUGUCAACAUUUUCCGUUCCGUUCAGUUUCGUUCGGUUUUCCGUUGGGUUUGAAGGGUCGCAGCUGCACGCAAAUAUUUGCCGCAUUUUUUAUGCGACCACAAAACAGUGGG ....((((((((.................(((((.((((..(((....))).)))).))))).(((((((..(((..((((....)))).))).......))))))).....)))))))) ( -37.60) >DroSec_CAF1 16138 110 - 1 GCUACCCGCUGUCAACAUUUUCCGUUCCGUUC----------CGUUUUCCGUGGGGUUUGAAGGGUCGCAGCUGCACUCAAAUAUUUGCCGCAUUUUUUAUGCGACCACAAAACAGUGGG ....((((((((......((((.(..(((...----------((.....)))))..)..))))(((((((((.(((..........))).))........))))))).....)))))))) ( -30.50) >DroSim_CAF1 16669 110 - 1 GCUACCCGCUGUCAACAUUUUCCGUUCCGUUC----------CGUUUUCCGUGGGGUUUGAAGGGUCGCAGCUGCACUCAAAUAUUUGCCGCAUUUUUUAUGCGACCACAAAACAGUGGG ....((((((((......((((.(..(((...----------((.....)))))..)..))))(((((((((.(((..........))).))........))))))).....)))))))) ( -30.50) >DroEre_CAF1 16937 93 - 1 GCUACCCACUGUCAACA---------------------------UUUUCCGAUGGGUUUGAAGGGUCGCAGCUGCACGCAAAUAUUUGCCGCAUUUUUUAUGCGACCACAAAACAGUGGG ....((((((((...((---------------------------((....))))..((((...(((((((..(((..((((....)))).))).......))))))).)))))))))))) ( -31.70) >DroYak_CAF1 18669 110 - 1 GCUACCCACUGUCAACAAUUUCCGUUCCGUUC----------UAUUUUCCGAUGGGUUUGAAGGGUCGCAGCUGCACGCAAAUAUUUGCCGCAUUUUUUAUGCGACCACAAAACAGUGGG ....((((((((......((((...((((((.----------........))))))...))))(((((((..(((..((((....)))).))).......))))))).....)))))))) ( -34.90) >consensus GCUACCCACUGUCAACAUUUUCCGUUCCGUUC__________CGUUUUCCGUUGGGUUUGAAGGGUCGCAGCUGCACGCAAAUAUUUGCCGCAUUUUUUAUGCGACCACAAAACAGUGGG ....((((((((....................................((....))((((...(((((((((.(((..........))).))........))))))).)))))))))))) (-28.78 = -28.54 + -0.24)


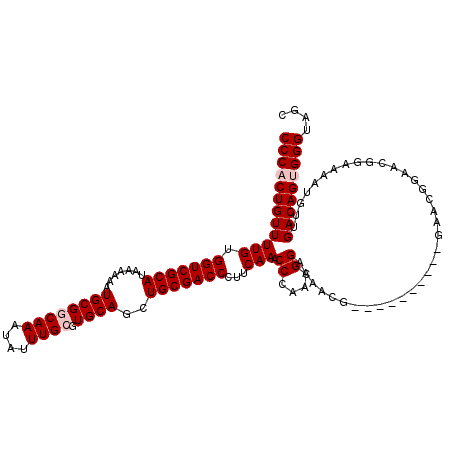
| Location | 21,045,866 – 21,045,979 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 81.50 |
| Mean single sequence MFE | -38.25 |
| Consensus MFE | -27.65 |
| Energy contribution | -29.15 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.620302 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21045866 113 + 22224390 CGAGGAUUCGCAACUGGCAAACAAAAAGUGGGGAAGUGGGAAGUGGUGGGUGGUGGUGGAAAGUGCUUUCCCG-AGGGGCUUAAGACCAUUUUCCAGGCCUCCCUUGCCUUGGU (((((....((.....)).......(((.((((...((((((((((((((.((((.(....).))))..)))(-((...)))...)))))))))))..)))).))).))))).. ( -36.50) >DroSec_CAF1 16248 111 + 1 CGAGGAUUCGCAACUGGGAAACCAAAUGGGGGGAUAUGGAAGGUGGUGGGUG---GUGGAAAGUGGUUUCCCAAAGGGGCUCAAGACCAUUUUCCAGGCCUCCUUGGCCUCGGG (((((.........(((....)))...((((((...(((((((((((((((.---(.(((((....))))))......))))...)))))))))))..))))))...))))).. ( -43.60) >DroSim_CAF1 16779 114 + 1 CGAGGAUUCGCAACUGGGAAACCAAAUGGGGGGAUAUGGAAGGUGGUGGGUGGUGGUGGAAAGUGGUUUCCCAAAGGGGCUUAAGACCAUUUUCCAGGCCUCCUUGGCCUCGGG (((((.(((.((..(((....)))..)).)))...............(((.(((..((((((((((((((((....)))....))))))))))))).))).)))...))))).. ( -44.20) >DroYak_CAF1 18779 105 + 1 CGAGGAUUGGGAACUGGCAAGUAAAAAGU-UGGAAAUGGGAGGUGGAAG------AUGGAAAGGGGUUUCCCGAGGG--CUUAAGGCCAUUUUCCAGGACUCCUUGGACUGGGG (.((.........(..((.........))-..)....(((((.((((((------((((....((((((.....)))--)))....))))))))))...)))))....)).).. ( -28.70) >consensus CGAGGAUUCGCAACUGGCAAACAAAAAGGGGGGAAAUGGAAGGUGGUGGGUG___GUGGAAAGUGGUUUCCCAAAGGGGCUUAAGACCAUUUUCCAGGCCUCCUUGGCCUCGGG (((((.........(((....)))...((((((...(((((((((((((((......(((((....))))).......))))...)))))))))))..))))))...))))).. (-27.65 = -29.15 + 1.50)

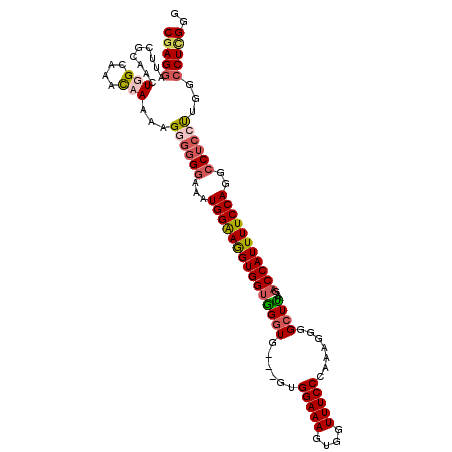

| Location | 21,045,906 – 21,046,003 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 77.73 |
| Mean single sequence MFE | -36.20 |
| Consensus MFE | -25.64 |
| Energy contribution | -26.82 |
| Covariance contribution | 1.19 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.71 |
| SVM decision value | 2.29 |
| SVM RNA-class probability | 0.991764 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21045906 97 + 22224390 AAGUGGUGGGUGGUGGUGGAAAGUGCUUUCCCG-AGGGGCUUAAGACCAUUUUCCAGGCCUCCCUUGCCUUGGUUAGGCCGAAAAGUGGAUGCC----------------AAAA ....((..((.((.((((((((((((((.(((.-..)))...)))..))))))))..))).))))..))(((((....(((.....)))..)))----------------)).. ( -32.80) >DroSec_CAF1 16288 94 + 1 AGGUGGUGGGUG---GUGGAAAGUGGUUUCCCAAAGGGGCUCAAGACCAUUUUCCAGGCCUCCUUGGCCUCGGGUAGGCC-AAAAAGCGAUGCC----------------AAAA .(((((.((((.---.((((((((((((((((....)))....))))))))))))).))))))(((((((.....)))))-))........)))----------------.... ( -39.20) >DroSim_CAF1 16819 98 + 1 AGGUGGUGGGUGGUGGUGGAAAGUGGUUUCCCAAAGGGGCUUAAGACCAUUUUCCAGGCCUCCUUGGCCUCGGGUAGGCCGAAAAAGCGAUGCC----------------AAAA .(((..((((.(((..((((((((((((((((....)))....))))))))))))).))).))(((((((.....))))))).....))..)))----------------.... ( -40.10) >DroYak_CAF1 18818 106 + 1 AGGUGGAAG------AUGGAAAGGGGUUUCCCGAGGG--CUUAAGGCCAUUUUCCAGGACUCCUUGGACUGGGGUAGGCCGAAAAAGCGAUGCCAAAAUACCAAAAUGCCAAAA .((((....------.(((...(((....)))....(--(((..((((...((((((..(.....)..))))))..))))....))))....)))...))))............ ( -32.70) >consensus AGGUGGUGGGUG___GUGGAAAGUGGUUUCCCAAAGGGGCUUAAGACCAUUUUCCAGGCCUCCUUGGCCUCGGGUAGGCCGAAAAAGCGAUGCC________________AAAA ....((((........((((((((((((((((....)))....)))))))))))))((((((((.......))).)))))..........)))).................... (-25.64 = -26.82 + 1.19)

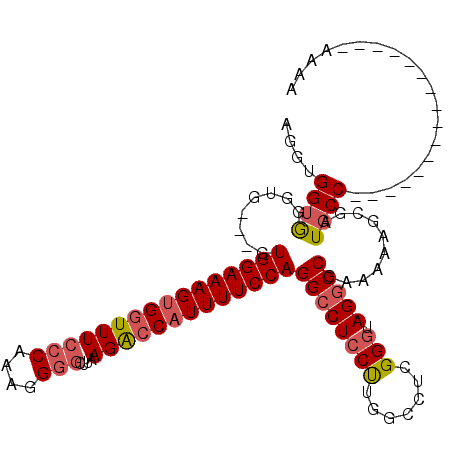
| Location | 21,045,906 – 21,046,003 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 77.73 |
| Mean single sequence MFE | -28.65 |
| Consensus MFE | -18.27 |
| Energy contribution | -20.40 |
| Covariance contribution | 2.13 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.922436 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21045906 97 - 22224390 UUUU----------------GGCAUCCACUUUUCGGCCUAACCAAGGCAAGGGAGGCCUGGAAAAUGGUCUUAAGCCCCU-CGGGAAAGCACUUUCCACCACCACCCACCACUU ...(----------------((.............((((.....))))..(((.((..((((((.((.(((..((...))-..)))...)).))))))...)).))).)))... ( -24.80) >DroSec_CAF1 16288 94 - 1 UUUU----------------GGCAUCGCUUUUU-GGCCUACCCGAGGCCAAGGAGGCCUGGAAAAUGGUCUUGAGCCCCUUUGGGAAACCACUUUCCAC---CACCCACCACCU ....----------------(((....((((((-(((((.....))))))))))))))((((((.((((......(((....)))..)))).)))))).---............ ( -32.40) >DroSim_CAF1 16819 98 - 1 UUUU----------------GGCAUCGCUUUUUCGGCCUACCCGAGGCCAAGGAGGCCUGGAAAAUGGUCUUAAGCCCCUUUGGGAAACCACUUUCCACCACCACCCACCACCU ...(----------------((....(((((((.(((((.....))))).))))))).((((((.((((......(((....)))..)))).)))))))))............. ( -31.00) >DroYak_CAF1 18818 106 - 1 UUUUGGCAUUUUGGUAUUUUGGCAUCGCUUUUUCGGCCUACCCCAGUCCAAGGAGUCCUGGAAAAUGGCCUUAAG--CCCUCGGGAAACCCCUUUCCAU------CUUCCACCU ..((((.(((..((((....(((............)))))))..)))))))((((...((((((..(((.....)--))...((....))..)))))).------.)))).... ( -26.40) >consensus UUUU________________GGCAUCGCUUUUUCGGCCUACCCGAGGCCAAGGAGGCCUGGAAAAUGGUCUUAAGCCCCUUCGGGAAACCACUUUCCAC___CACCCACCACCU ....................((....(((((((.(((((.....))))).))))))).((((((.((((((((((....)))))))..))).))))))..........)).... (-18.27 = -20.40 + 2.13)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:05:18 2006