
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 21,043,864 – 21,044,012 |
| Length | 148 |
| Max. P | 0.860364 |

| Location | 21,043,864 – 21,043,981 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.75 |
| Mean single sequence MFE | -33.91 |
| Consensus MFE | -27.51 |
| Energy contribution | -27.97 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.860364 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
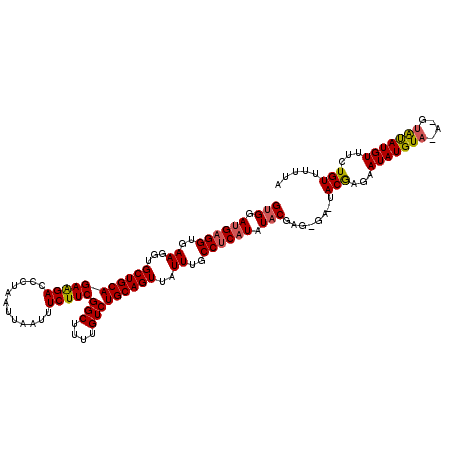
>X_DroMel_CAF1 21043864 117 - 22224390 GUGGAUGAGGUGAAGGUGCUGCAGAAGACCCUAAUUAAUUUCUUCGGCUUUUGUCUGCAGUUAUUUGCCUCAUAUACGAG-GAGGACAAGUAUAUGUACA--UAUAUGUUUCUGUAUUUA (((.((((((..((...(((((((((((............)))))(((....)))))))))..))..)))))).)))..(-((((....((((((....)--))))).)))))....... ( -35.60) >DroSim_CAF1 14814 116 - 1 GUGGAUGAGGUGAAGGUGCUGCAGAAGACCCUAAUUAAUUUCUUCGGCUUUUGUCUGCAGUUAUUUGCCUCAUAUACGAG----UACGAGCAUAUGUAUAUGUAUAUGUUUCUGUUUUUA (((.((((((..((...(((((((((((............)))))(((....)))))))))..))..)))))).)))...----...(((((((((......)))))))))......... ( -36.10) >DroYak_CAF1 16257 116 - 1 GCGAACGAGGUGAAGGUGCUGCAGAGGACCCUAAUUAAUUUCUUCGGCUUUUGUCUGCAGUUAUUUUCCGCAUAUACACGAGAAUACGAGAAUAUGU----GUGCAUGUUUCUGUUUGUA ((((((((..(((((((((((((((.((.((..............))...)).))))))).)))))))(((((((.(.((......)).).))))))----).....)..)).)))))). ( -30.04) >consensus GUGGAUGAGGUGAAGGUGCUGCAGAAGACCCUAAUUAAUUUCUUCGGCUUUUGUCUGCAGUUAUUUGCCUCAUAUACGAG_GA_UACGAGAAUAUGUA_A_GUAUAUGUUUCUGUUUUUA (((.((((((..((...(((((((((((............)))))(((....)))))))))..))..)))))).)))........(((...(((((((....)))))))...)))..... (-27.51 = -27.97 + 0.45)

| Location | 21,043,901 – 21,044,012 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.27 |
| Mean single sequence MFE | -34.77 |
| Consensus MFE | -28.28 |
| Energy contribution | -29.64 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.859813 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21043901 111 - 22224390 UAAUGAUGGUGAUGAUAUGGCCAUGAUG---------GCGGUGGAUGAGGUGAAGGUGCUGCAGAAGACCCUAAUUAAUUUCUUCGGCUUUUGUCUGCAGUUAUUUGCCUCAUAUACGAG ..((.(((((.((...)).))))).)).---------...(((.((((((..((...(((((((((((............)))))(((....)))))))))..))..)))))).)))... ( -36.50) >DroSec_CAF1 14328 111 - 1 UAAUGAUGGUGAUGAUAUGGCCAUGAUG---------GCGGUGGAUGAGGUGAAGGUGCUGCAGAAGACCCAAAUUAAUUUCUUCGGCUUUUGUCUGCAGUUAUUUGCCUCAUAUGCGAG ..((.(((((.((...)).))))).)).---------(((....((((((..((...(((((((((((............)))))(((....)))))))))..))..)))))).)))... ( -37.30) >DroSim_CAF1 14850 111 - 1 UAAUGAUGGUGAUGAUAUGGCCAUGAUG---------GCGGUGGAUGAGGUGAAGGUGCUGCAGAAGACCCUAAUUAAUUUCUUCGGCUUUUGUCUGCAGUUAUUUGCCUCAUAUACGAG ..((.(((((.((...)).))))).)).---------...(((.((((((..((...(((((((((((............)))))(((....)))))))))..))..)))))).)))... ( -36.50) >DroEre_CAF1 15040 120 - 1 UAAUGAUGGUGAUGAUAUGGCCAUGAUGGCGGUAAUGGCGGCGAACGAGGUGAAGGUGCUGCAGAAGACCCUAAUUAAUUUCUUCGCCUUUUGUCUGCAGUUAUUUGCCGCAUAUACGAG ..((.(((((.((...)).))))).)).(((((((((((((((((.(((((..(((..((.....))..))).....)))))))))))...((....)))))).)))))))......... ( -35.80) >DroYak_CAF1 16293 111 - 1 UAAUGUUGGUGAUGAUAUGGCCACGAUG---------UCGGCGAACGAGGUGAAGGUGCUGCAGAGGACCCUAAUUAAUUUCUUCGGCUUUUGUCUGCAGUUAUUUUCCGCAUAUACACG ........(((...((((((((......---------..)))......((.(((...((((((((.((.((..............))...)).))))))))..))).)).))))).))). ( -27.74) >consensus UAAUGAUGGUGAUGAUAUGGCCAUGAUG_________GCGGUGGAUGAGGUGAAGGUGCUGCAGAAGACCCUAAUUAAUUUCUUCGGCUUUUGUCUGCAGUUAUUUGCCUCAUAUACGAG ..((.(((((.((...)).))))).)).............(((.((((((..((...(((((((((((............)))))(((....)))))))))..))..)))))).)))... (-28.28 = -29.64 + 1.36)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:05:11 2006