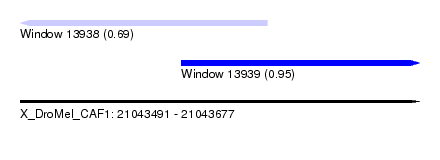
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 21,043,491 – 21,043,677 |
| Length | 186 |
| Max. P | 0.954275 |

| Location | 21,043,491 – 21,043,606 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.19 |
| Mean single sequence MFE | -42.74 |
| Consensus MFE | -30.67 |
| Energy contribution | -30.07 |
| Covariance contribution | -0.60 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.690657 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21043491 115 - 22224390 GGCCUCCUUCUCCUGUUCGGCCAACUUCUCGAUAAGCGUUUAAUUGCUGGUCGAGGACCCAAUGU----CCCAUCUUCUGGGAUGCCUGGGAUUUUGGGUGGCCCAGAACG-GUCUGUUA ............(((((((((((..((((((((.((((......)))).))))))))(((((.((----(((((((....)))....)))))).))))))))))..)))))-)....... ( -46.00) >DroSec_CAF1 13931 113 - 1 GGCCUCCUUCUCCUCUUCGGCCAACUUCUCGAUAAGCGUUUAAUUGCUGGUCGAGGACCCAACGU----CCCGUCUUCUGGGAUGCCCG--AUAUUGGGUGGCACAGAACG-GGCUGUUG ((((..............))))...((((((((.((((......)))).))))))))..(((((.----(((((..((((...((((((--....))))))...)))))))-)).))))) ( -44.94) >DroSim_CAF1 14449 114 - 1 GGCCUCCUUCUCCUCUUCGGCCAACUUCUCGAUAAGCGUUUAAUUGCUGGUCGAGGACCCAACGU----CCCGUCUUCUGGGAUGCCCG--AUAUUGAGUGGCACAGAACGAGGCUGUUG ((((((.((((((((.((((.....((((((((.((((......)))).)))))))).....(((----((((.....))))))).)))--)....))).)....)))).)))))).... ( -42.60) >DroEre_CAF1 14679 115 - 1 GGCCUCCUUCUCCUCUUCGGCCAACUUCUCGAUAAGCGUUUAAUUGCUGGUCGAGGACCCAACGUCCGUCCUGUCUUCUGGGAUGCCCG--AUUGUGGGUGGUG---AACUGGGCGGUUG .((((..(((.((.............(((((((.((((......)))).)))))))(((((.((..((((((.......))))))..))--....))))))).)---))..))))..... ( -41.80) >DroYak_CAF1 15886 109 - 1 GGCCUCCUUCUCCUCUUUGGCCAACUUCUCGAUAAGCGUUUAAUUGCUGGUCGAGGACCCAACGU----CCUGUCUUCUGGGAUGCCCG--AUUGUGGGUGGUU-----CGGGGCUGUGA ((((..............))))...((((((((.((((......)))).))))))))((((((((----(((.......)))))(((((--....))))).)))-----.)))....... ( -38.34) >consensus GGCCUCCUUCUCCUCUUCGGCCAACUUCUCGAUAAGCGUUUAAUUGCUGGUCGAGGACCCAACGU____CCCGUCUUCUGGGAUGCCCG__AUUUUGGGUGGCACAGAACG_GGCUGUUG .(((.(((..........(((....((((((((.((((......)))).))))))))(((((((.......)))....))))..))).........))).)))................. (-30.67 = -30.07 + -0.60)


| Location | 21,043,566 – 21,043,677 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.23 |
| Mean single sequence MFE | -34.61 |
| Consensus MFE | -29.78 |
| Energy contribution | -30.46 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.954275 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21043566 111 + 22224390 AACGCUUAUCGAGAAGUUGGCCGAACAGGAGAAGGAGGCCGGCCAGCGGAGAG---------GCAGGGCAGAAAACAGACGACAAUGAGCUGCCUUGCAUAUAAUUGAGGUUAUGGCCGA ........((((...((((((((..(..........)..))))))))......---------(((((((((....((........))..)))))))))......))))(((....))).. ( -36.20) >DroSec_CAF1 14004 107 + 1 AACGCUUAUCGAGAAGUUGGCCGAAGAGGAGAAGGAGGCCGGCCAGCGCAGAG---------GCAGGGCGGAAAACAGACGACAAUGAGC----UUGCAUAUAAUUGAGGUUAUGGCCGA ..(((((........((((((((................))))))))((....---------)).))))).........((.((...(((----((.((......))))))).))..)). ( -25.99) >DroSim_CAF1 14523 111 + 1 AACGCUUAUCGAGAAGUUGGCCGAAGAGGAGAAGGAGGCCGGCCAGCGCAGAG---------GCAGGGCGGAAAACAGACGACAAUGAGCUGCCUUGCAUAUAAUUGAGGUUAUGGCCGA ........((((...((((((((................))))))))......---------(((((((((....((........))..)))))))))......))))(((....))).. ( -34.19) >DroEre_CAF1 14754 119 + 1 AACGCUUAUCGAGAAGUUGGCCGAAGAGGAGAAGGAGGCCGGCCAGGGCAG-GCAGUGCAGUGCAGGGCAGAAAACAGACGACAAUGAGCUGCCUUGCAUAUAAUUGAGGUUAUGGCCGA ...((((......)))).((((..............))))(((((..((..-.((((...(((((((((((....((........))..)))))))))))...))))..))..))))).. ( -43.74) >DroYak_CAF1 15955 110 + 1 AACGCUUAUCGAGAAGUUGGCCAAAGAGGAGAAGGAGGCCGGCCAGGGUGG-G---------GCAGGGCAGAAAACAGACGACAAUGAGCUGCCUUGCAUAUAAUUGAGGUUAUGGCCGA ....((((((.....(((((((..............)))))))...)))))-)---------(((((((((....((........))..)))))))))..........(((....))).. ( -32.94) >consensus AACGCUUAUCGAGAAGUUGGCCGAAGAGGAGAAGGAGGCCGGCCAGCGCAGAG_________GCAGGGCAGAAAACAGACGACAAUGAGCUGCCUUGCAUAUAAUUGAGGUUAUGGCCGA ...((((......)))).((((..............))))((((..................(((((((((....((........))..)))))))))..(((((....))))))))).. (-29.78 = -30.46 + 0.68)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:05:09 2006