
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 21,043,147 – 21,043,458 |
| Length | 311 |
| Max. P | 0.999970 |

| Location | 21,043,147 – 21,043,267 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.42 |
| Mean single sequence MFE | -30.38 |
| Consensus MFE | -26.74 |
| Energy contribution | -26.74 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.967618 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21043147 120 + 22224390 GUAUCAACAAUACCACCGCUGUUGCUGCAGCACCACCAACUGAAAGCACUGAAAUCCACUACAAUGGAGGCAUUUAAUAGCAGCCCGAUGCACUUUCAUUGGAGUUUCCACUCCGAGGGC .................(((((....)))))....((...((((((...((...((((......))))..)).......(((......))).))))))(((((((....))))))))).. ( -29.50) >DroSec_CAF1 13576 120 + 1 GUAUCACCAAUACCACCCCAGCUGGUGCAGCACCACCAACUAAAAGCACUGGAAUCCACUACAAUGGCGGCAUUUAAUAGCAGCCCGGUGCACUUUUAUUGGAGUUUCCACUCCAAGUGC ....(((((.............)))))..((((............(((((((...(((......)))..((........))...))))))).......(((((((....))))))))))) ( -31.12) >DroSim_CAF1 14094 120 + 1 GUAUCACCAAUACCACCCCAGCUGGUGCAGCACCACCAACUAAAAGCACUGGAAUCCACUACAAUGGCGGCAUUUAAUAGCAGCCCGGUGCACUUUCAUUGGAGUUUCCACUCCAAGUGC ....(((((.............)))))..((((............(((((((...(((......)))..((........))...))))))).......(((((((....))))))))))) ( -31.12) >DroEre_CAF1 14328 120 + 1 GUAUCACCAAUACCCGUCCAGCUCCGGCAGCACAACCAACUAAAAGCACUGAAAUCCACUACAAUGGCGGCAUUCAAUAGCAGCCCGGUGCAUUUUCAUUGGAGUUUCCACUCCAAGUGC .............(((........)))..((((............((((((..............(((.((........)).))))))))).......(((((((....))))))))))) ( -28.53) >DroYak_CAF1 15536 120 + 1 GUAUCACCAAUACCAGCUCAGUUGCUGCAGCACAACCAACUAAAAGCACUGAAAUCCACUACAAUGGCGGCAUUCACUAGCAGCCCGGUGCAUUUUUAUUGGAGUUUCCACUCCAAGUGC .............((((......))))..((((............((((((..............(((.((........)).))))))))).......(((((((....))))))))))) ( -31.63) >consensus GUAUCACCAAUACCACCCCAGCUGCUGCAGCACCACCAACUAAAAGCACUGAAAUCCACUACAAUGGCGGCAUUUAAUAGCAGCCCGGUGCACUUUCAUUGGAGUUUCCACUCCAAGUGC .............((((......))))..((((............((((((....(((......))).(((...........))))))))).......(((((((....))))))))))) (-26.74 = -26.74 + 0.00)


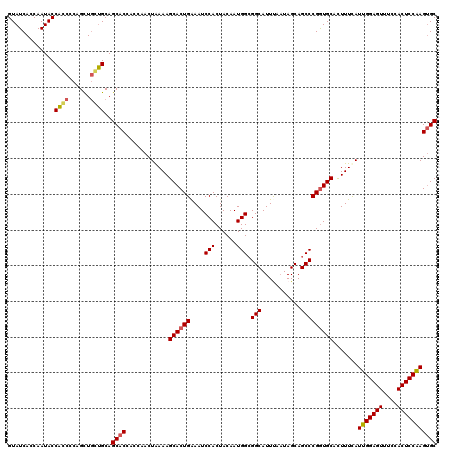
| Location | 21,043,147 – 21,043,267 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.42 |
| Mean single sequence MFE | -43.84 |
| Consensus MFE | -32.98 |
| Energy contribution | -34.14 |
| Covariance contribution | 1.16 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.923024 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21043147 120 - 22224390 GCCCUCGGAGUGGAAACUCCAAUGAAAGUGCAUCGGGCUGCUAUUAAAUGCCUCCAUUGUAGUGGAUUUCAGUGCUUUCAGUUGGUGGUGCUGCAGCAACAGCGGUGGUAUUGUUGAUAC ......(((((....)))))..((((((..(....(((...........)))(((((....))))).....)..))))))((..(..((((..(.((....)).)..))))..)..)).. ( -43.70) >DroSec_CAF1 13576 120 - 1 GCACUUGGAGUGGAAACUCCAAUAAAAGUGCACCGGGCUGCUAUUAAAUGCCGCCAUUGUAGUGGAUUCCAGUGCUUUUAGUUGGUGGUGCUGCACCAGCUGGGGUGGUAUUGGUGAUAC (((((((((((....)))))).....)))))(((((....))....(((((((((...(((.(((...))).)))..((((((((((......))))))))))))))))))))))..... ( -48.70) >DroSim_CAF1 14094 120 - 1 GCACUUGGAGUGGAAACUCCAAUGAAAGUGCACCGGGCUGCUAUUAAAUGCCGCCAUUGUAGUGGAUUCCAGUGCUUUUAGUUGGUGGUGCUGCACCAGCUGGGGUGGUAUUGGUGAUAC (((((((((((....)))))).....)))))(((((....))....(((((((((...(((.(((...))).)))..((((((((((......))))))))))))))))))))))..... ( -48.70) >DroEre_CAF1 14328 120 - 1 GCACUUGGAGUGGAAACUCCAAUGAAAAUGCACCGGGCUGCUAUUGAAUGCCGCCAUUGUAGUGGAUUUCAGUGCUUUUAGUUGGUUGUGCUGCCGGAGCUGGACGGGUAUUGGUGAUAC .((((((((((....)))))).....(((((.(((....((.((((((..((((.......))))..)))))))).((((((((((......))))..)))))))))))))))))).... ( -38.70) >DroYak_CAF1 15536 120 - 1 GCACUUGGAGUGGAAACUCCAAUAAAAAUGCACCGGGCUGCUAGUGAAUGCCGCCAUUGUAGUGGAUUUCAGUGCUUUUAGUUGGUUGUGCUGCAGCAACUGAGCUGGUAUUGGUGAUAC .((((((((((....)))))))......((((...(((.((........)).)))..)))))))....((((((((((((((((.(((.....)))))))))))..))))))))...... ( -39.40) >consensus GCACUUGGAGUGGAAACUCCAAUGAAAGUGCACCGGGCUGCUAUUAAAUGCCGCCAUUGUAGUGGAUUUCAGUGCUUUUAGUUGGUGGUGCUGCAGCAGCUGGGGUGGUAUUGGUGAUAC .((((((((((....)))))))......((((...(((.((........)).)))..)))))))....((((((((((((((((.((......)).))))))))..))))))))...... (-32.98 = -34.14 + 1.16)

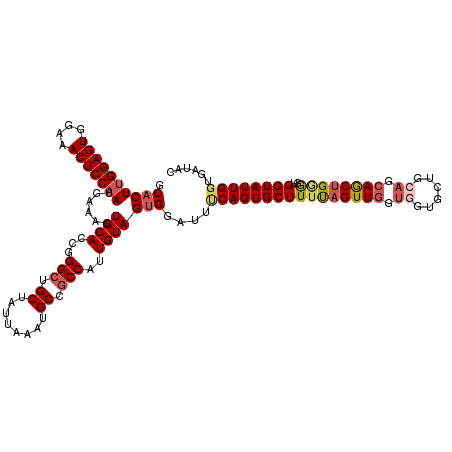
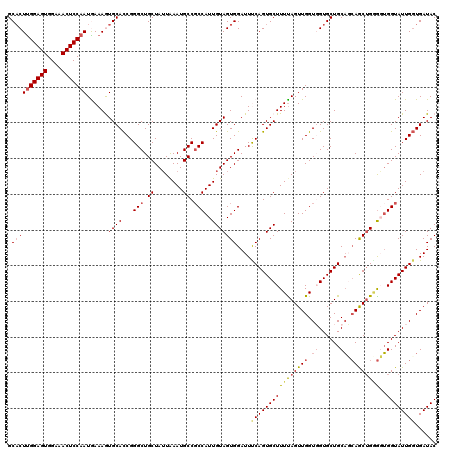
| Location | 21,043,187 – 21,043,307 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.33 |
| Mean single sequence MFE | -40.10 |
| Consensus MFE | -39.14 |
| Energy contribution | -38.98 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.98 |
| SVM decision value | 3.30 |
| SVM RNA-class probability | 0.998954 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21043187 120 + 22224390 UGAAAGCACUGAAAUCCACUACAAUGGAGGCAUUUAAUAGCAGCCCGAUGCACUUUCAUUGGAGUUUCCACUCCGAGGGCCGGCGAUCCGAUCGGAUCGGGCAUCAAUUUCGUGCUGUGC ....(((((.((((((((......)))).((........)).(((((((.(.......(((((((....))))))).(..(((....)))..)).))))))).....))))))))).... ( -40.50) >DroSec_CAF1 13616 120 + 1 UAAAAGCACUGGAAUCCACUACAAUGGCGGCAUUUAAUAGCAGCCCGGUGCACUUUUAUUGGAGUUUCCACUCCAAGUGCCGGCGAUCCGAUCGGAUCGGGCAUCAAUUUCGUGCUGUGC .....((((...........((...(((.((........)).)))..))((((.....(((((((....)))))))(((((..((((((....))))))))))).......)))).)))) ( -40.70) >DroSim_CAF1 14134 120 + 1 UAAAAGCACUGGAAUCCACUACAAUGGCGGCAUUUAAUAGCAGCCCGGUGCACUUUCAUUGGAGUUUCCACUCCAAGUGCCGGCGAUCCGAUCGGAUCGGGCAUCAAUUUCGUGCUGUGC .....((((...........((...(((.((........)).)))..))((((.....(((((((....)))))))(((((..((((((....))))))))))).......)))).)))) ( -40.70) >DroEre_CAF1 14368 120 + 1 UAAAAGCACUGAAAUCCACUACAAUGGCGGCAUUCAAUAGCAGCCCGGUGCAUUUUCAUUGGAGUUUCCACUCCAAGUGCCGGCGAUCCGAUCAGAUCGGGCAUCAAUUUCGUGCUGUGC ....((((((((((..((((.....(((.((........)).))).))))...)))))(((((((....)))))))(((((..(((((......)))))))))).......))))).... ( -37.60) >DroYak_CAF1 15576 120 + 1 UAAAAGCACUGAAAUCCACUACAAUGGCGGCAUUCACUAGCAGCCCGGUGCAUUUUUAUUGGAGUUUCCACUCCAAGUGCCGGCGAUCCGAUCGGAUCGGGCAUCAAUUUCGUGCUGUCC ....(((((.(((((..........(((.((........)).))).(((((.......(((((((....))))))).......((((((....)))))).))))).)))))))))).... ( -41.00) >consensus UAAAAGCACUGAAAUCCACUACAAUGGCGGCAUUUAAUAGCAGCCCGGUGCACUUUCAUUGGAGUUUCCACUCCAAGUGCCGGCGAUCCGAUCGGAUCGGGCAUCAAUUUCGUGCUGUGC ....(((((.(((((..........(((.((........)).))).(((((.......(((((((....))))))).......((((((....)))))).))))).)))))))))).... (-39.14 = -38.98 + -0.16)

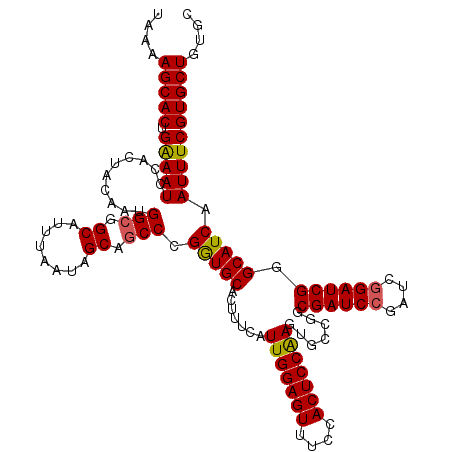

| Location | 21,043,187 – 21,043,307 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.33 |
| Mean single sequence MFE | -45.36 |
| Consensus MFE | -42.82 |
| Energy contribution | -43.46 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.00 |
| Structure conservation index | 0.94 |
| SVM decision value | 5.04 |
| SVM RNA-class probability | 0.999970 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21043187 120 - 22224390 GCACAGCACGAAAUUGAUGCCCGAUCCGAUCGGAUCGCCGGCCCUCGGAGUGGAAACUCCAAUGAAAGUGCAUCGGGCUGCUAUUAAAUGCCUCCAUUGUAGUGGAUUUCAGUGCUUUCA ((((.((((......((.(((((((((....))))))..)))..))(((((....))))).......))))....(((...........)))(((((....))))).....))))..... ( -42.70) >DroSec_CAF1 13616 120 - 1 GCACAGCACGAAAUUGAUGCCCGAUCCGAUCGGAUCGCCGGCACUUGGAGUGGAAACUCCAAUAAAAGUGCACCGGGCUGCUAUUAAAUGCCGCCAUUGUAGUGGAUUCCAGUGCUUUUA ((((.((((........((((((((((....))))))..)))).(((((((....))))))).....))))...((.(..((((..((((....))))))))..)...)).))))..... ( -46.20) >DroSim_CAF1 14134 120 - 1 GCACAGCACGAAAUUGAUGCCCGAUCCGAUCGGAUCGCCGGCACUUGGAGUGGAAACUCCAAUGAAAGUGCACCGGGCUGCUAUUAAAUGCCGCCAUUGUAGUGGAUUCCAGUGCUUUUA ((((.((((........((((((((((....))))))..)))).(((((((....))))))).....))))...((.(..((((..((((....))))))))..)...)).))))..... ( -46.20) >DroEre_CAF1 14368 120 - 1 GCACAGCACGAAAUUGAUGCCCGAUCUGAUCGGAUCGCCGGCACUUGGAGUGGAAACUCCAAUGAAAAUGCACCGGGCUGCUAUUGAAUGCCGCCAUUGUAGUGGAUUUCAGUGCUUUUA ....(((((((((((.(((((((((((....))))))..)))..(((((((....)))))))......((((...(((.((........)).)))..)))))).)))))).))))).... ( -44.70) >DroYak_CAF1 15576 120 - 1 GGACAGCACGAAAUUGAUGCCCGAUCCGAUCGGAUCGCCGGCACUUGGAGUGGAAACUCCAAUAAAAAUGCACCGGGCUGCUAGUGAAUGCCGCCAUUGUAGUGGAUUUCAGUGCUUUUA .((.(((((((((((.(((((((((((....))))))..)))..(((((((....)))))))......((((...(((.((........)).)))..)))))).)))))).))))).)). ( -47.00) >consensus GCACAGCACGAAAUUGAUGCCCGAUCCGAUCGGAUCGCCGGCACUUGGAGUGGAAACUCCAAUGAAAGUGCACCGGGCUGCUAUUAAAUGCCGCCAUUGUAGUGGAUUUCAGUGCUUUUA ....(((((((((((.(((((((((((....))))))..)))..(((((((....)))))))......((((...(((.((........)).)))..)))))).)))))).))))).... (-42.82 = -43.46 + 0.64)



| Location | 21,043,227 – 21,043,347 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.00 |
| Mean single sequence MFE | -42.09 |
| Consensus MFE | -39.92 |
| Energy contribution | -39.80 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.95 |
| SVM decision value | 2.92 |
| SVM RNA-class probability | 0.997734 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21043227 120 + 22224390 CAGCCCGAUGCACUUUCAUUGGAGUUUCCACUCCGAGGGCCGGCGAUCCGAUCGGAUCGGGCAUCAAUUUCGUGCUGUGCCUGACAGCGGCUGUCGCUUAUAAAUUGCAGAUUCCACAAA (((((.(((((.......(((((((....))))))).......((((((....)))))).)))))........(((((.....))))))))))..((.........))............ ( -41.00) >DroSec_CAF1 13656 120 + 1 CAGCCCGGUGCACUUUUAUUGGAGUUUCCACUCCAAGUGCCGGCGAUCCGAUCGGAUCGGGCAUCAAUUUCGUGCUGUGCCUGACAGCGGCUGUCGCUUAUAAAUUGCAGAUUCCACAAA ..((((((..(.......(((((((....))))))))..))...(((((....))))))))).........(((((((.....)))))((..(((((.........)).))).))))... ( -40.81) >DroSim_CAF1 14174 120 + 1 CAGCCCGGUGCACUUUCAUUGGAGUUUCCACUCCAAGUGCCGGCGAUCCGAUCGGAUCGGGCAUCAAUUUCGUGCUGUGCCUGACAGCGGCUGUCGCUUAUAAAUUGCAGAUUCCACAAA ..((((((..(.......(((((((....))))))))..))...(((((....))))))))).........(((((((.....)))))((..(((((.........)).))).))))... ( -40.81) >DroEre_CAF1 14408 120 + 1 CAGCCCGGUGCAUUUUCAUUGGAGUUUCCACUCCAAGUGCCGGCGAUCCGAUCAGAUCGGGCAUCAAUUUCGUGCUGUGCCUGACAGCGGCUGUCGCUUAUAAAUUGCAGAGCCCACAAA ..((((((..(.......(((((((....))))))))..))...((((......)))))))).........(((((((.....)))))((((.(.((.........))).)))).))... ( -39.71) >DroYak_CAF1 15616 120 + 1 CAGCCCGGUGCAUUUUUAUUGGAGUUUCCACUCCAAGUGCCGGCGAUCCGAUCGGAUCGGGCAUCAAUUUCGUGCUGUCCCUGACAGCGGCUGUCGCUUAUAAAUUGCAGUGCCCACAAA ..(((.((..(.......(((((((....))))))))..)))))(((((....)))))((((((((((((...((((((...))))))(((....)))...))))))..))))))..... ( -48.11) >consensus CAGCCCGGUGCACUUUCAUUGGAGUUUCCACUCCAAGUGCCGGCGAUCCGAUCGGAUCGGGCAUCAAUUUCGUGCUGUGCCUGACAGCGGCUGUCGCUUAUAAAUUGCAGAUUCCACAAA (((((.(((((.......(((((((....))))))).......((((((....)))))).)))))........(((((.....))))))))))..((.........))............ (-39.92 = -39.80 + -0.12)

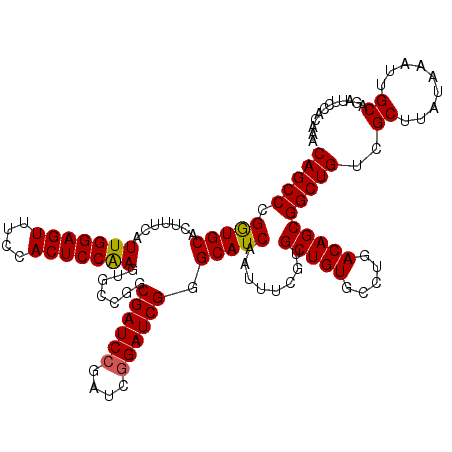
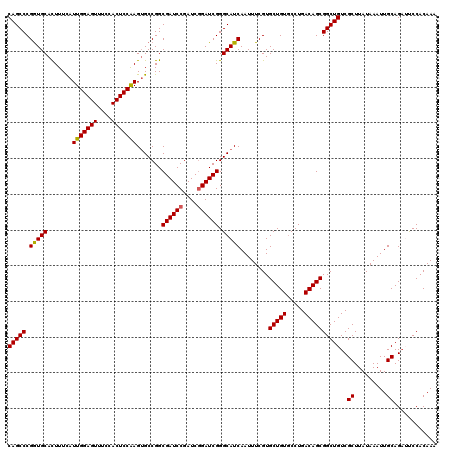
| Location | 21,043,227 – 21,043,347 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.00 |
| Mean single sequence MFE | -44.10 |
| Consensus MFE | -40.96 |
| Energy contribution | -41.20 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.93 |
| SVM decision value | 3.39 |
| SVM RNA-class probability | 0.999132 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21043227 120 - 22224390 UUUGUGGAAUCUGCAAUUUAUAAGCGACAGCCGCUGUCAGGCACAGCACGAAAUUGAUGCCCGAUCCGAUCGGAUCGCCGGCCCUCGGAGUGGAAACUCCAAUGAAAGUGCAUCGGGCUG .(((..(...)..)))...........((((((((....)))...((((......((.(((((((((....))))))..)))..))(((((....))))).......))))....))))) ( -42.40) >DroSec_CAF1 13656 120 - 1 UUUGUGGAAUCUGCAAUUUAUAAGCGACAGCCGCUGUCAGGCACAGCACGAAAUUGAUGCCCGAUCCGAUCGGAUCGCCGGCACUUGGAGUGGAAACUCCAAUAAAAGUGCACCGGGCUG .(((..(...)..)))...........((((((((....)))...((((........((((((((((....))))))..)))).(((((((....))))))).....))))....))))) ( -44.90) >DroSim_CAF1 14174 120 - 1 UUUGUGGAAUCUGCAAUUUAUAAGCGACAGCCGCUGUCAGGCACAGCACGAAAUUGAUGCCCGAUCCGAUCGGAUCGCCGGCACUUGGAGUGGAAACUCCAAUGAAAGUGCACCGGGCUG .(((..(...)..)))...........((((((((....)))...((((........((((((((((....))))))..)))).(((((((....))))))).....))))....))))) ( -44.90) >DroEre_CAF1 14408 120 - 1 UUUGUGGGCUCUGCAAUUUAUAAGCGACAGCCGCUGUCAGGCACAGCACGAAAUUGAUGCCCGAUCUGAUCGGAUCGCCGGCACUUGGAGUGGAAACUCCAAUGAAAAUGCACCGGGCUG (((((.((((.(((.........)))..))))(((((.....))))))))))......(((((((((....)))))((...((.(((((((....))))))))).....))...)))).. ( -42.10) >DroYak_CAF1 15616 120 - 1 UUUGUGGGCACUGCAAUUUAUAAGCGACAGCCGCUGUCAGGGACAGCACGAAAUUGAUGCCCGAUCCGAUCGGAUCGCCGGCACUUGGAGUGGAAACUCCAAUAAAAAUGCACCGGGCUG .....(((((...((((((....((....)).((((((...))))))...)))))).)))))(((((....)))))(((.(((.(((((((....)))))))......)))....))).. ( -46.20) >consensus UUUGUGGAAUCUGCAAUUUAUAAGCGACAGCCGCUGUCAGGCACAGCACGAAAUUGAUGCCCGAUCCGAUCGGAUCGCCGGCACUUGGAGUGGAAACUCCAAUGAAAGUGCACCGGGCUG .(((..(...)..)))...........((((((((((.....))))).((.......((((((((((....))))))..)))).(((((((....)))))))...........))))))) (-40.96 = -41.20 + 0.24)


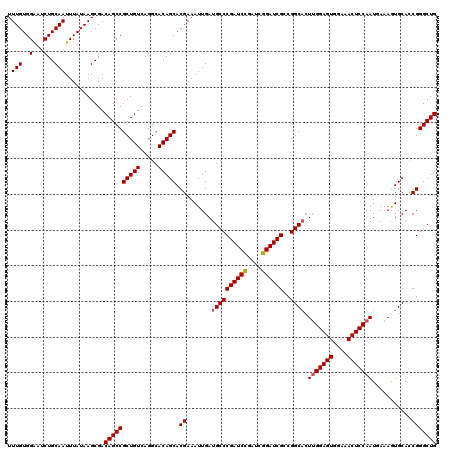
| Location | 21,043,307 – 21,043,418 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.59 |
| Mean single sequence MFE | -37.21 |
| Consensus MFE | -29.07 |
| Energy contribution | -29.87 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.947953 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21043307 111 + 22224390 CUGACAGCGGCUGUCGCUUAUAAAUUGCAGAUUCCACAAAAACAGCAAGAACAACAAGGAC--CAUGCGC-------CAUGGCCACUGUGGCAACAACGGCGACAAGCGCCCUGACAUUU ....(((.((((((((((........(((...(((......................))).--..)))((-------(((((...)))))))......)))))))...))))))...... ( -31.25) >DroSec_CAF1 13736 118 + 1 CUGACAGCGGCUGUCGCUUAUAAAUUGCAGAUUCCACAAAAACAGCAAGAACAACAAGGGC--CAUGGGCCAUGGGCCAUGGCCACUGUGGCAACAACGGCGACAAGCGCCCUGACAUUU ....(((.((((((((((......((((................)))).....(((..(((--(((((.(....).))))))))..)))(....)...)))))))...))))))...... ( -42.19) >DroSim_CAF1 14254 118 + 1 CUGACAGCGGCUGUCGCUUAUAAAUUGCAGAUUCCACAAAAACAGCAAGAACAACAAGGGC--CAUGGGCCAUGGGCCAUUGCCACUGUGGCAACAACGGCGACAAGCGCCCUGACAUUU ....(((.((((((((((......((((................)))).........((.(--((((...))))).)).(((((.....)))))....)))))))...))))))...... ( -35.69) >DroEre_CAF1 14488 118 + 1 CUGACAGCGGCUGUCGCUUAUAAAUUGCAGAGCCCACAAAAACAGCAAGAACAACAAGGGC--CAUGGGCCAUGGGCCAUGGCCACUAUGGCAACAACGGCGACAAGCGCCCUGACAUUU ....(((.((((((((((......((((...((...........))........(((((((--(((((.(....).)))))))).)).))))))....)))))))...))))))...... ( -41.80) >DroYak_CAF1 15696 120 + 1 CUGACAGCGGCUGUCGCUUAUAAAUUGCAGUGCCCACAAAAACAGCAAAAACAGCAAGAACAACAAGGGCCAUGGGCCAUGGCCACUAUGGCAACAACGGCGACAAGCGCCCUGACAUUU ....(((.((((((((((......(((...((((..........((.......))..........(((((((((...))))))).))..)))).))).)))))))...))))))...... ( -35.10) >consensus CUGACAGCGGCUGUCGCUUAUAAAUUGCAGAUUCCACAAAAACAGCAAGAACAACAAGGGC__CAUGGGCCAUGGGCCAUGGCCACUGUGGCAACAACGGCGACAAGCGCCCUGACAUUU ....(((.((((((((((......((((................))))...............(((((.(....).)))))(((.....)))......)))))))...))))))...... (-29.07 = -29.87 + 0.80)



| Location | 21,043,307 – 21,043,418 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.59 |
| Mean single sequence MFE | -41.16 |
| Consensus MFE | -32.44 |
| Energy contribution | -32.80 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.620249 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21043307 111 - 22224390 AAAUGUCAGGGCGCUUGUCGCCGUUGUUGCCACAGUGGCCAUG-------GCGCAUG--GUCCUUGUUGUUCUUGCUGUUUUUGUGGAAUCUGCAAUUUAUAAGCGACAGCCGCUGUCAG ...((.((((((.(.((.((((((.((..(....)..)).)))-------))))).)--((((((((.....((((.((((.....))))..))))...))))).))).))).))).)). ( -35.90) >DroSec_CAF1 13736 118 - 1 AAAUGUCAGGGCGCUUGUCGCCGUUGUUGCCACAGUGGCCAUGGCCCAUGGCCCAUG--GCCCUUGUUGUUCUUGCUGUUUUUGUGGAAUCUGCAAUUUAUAAGCGACAGCCGCUGUCAG ...((.(((.(.(((.(((((.((....((.((((.((((((((((...)).)))))--))).)))).))....))(((..(((..(...)..)))...))).))))))))).))).)). ( -44.30) >DroSim_CAF1 14254 118 - 1 AAAUGUCAGGGCGCUUGUCGCCGUUGUUGCCACAGUGGCAAUGGCCCAUGGCCCAUG--GCCCUUGUUGUUCUUGCUGUUUUUGUGGAAUCUGCAAUUUAUAAGCGACAGCCGCUGUCAG ...((.(((.(.(((.(((((.(((((..((((((..((((..((.((.((((...)--)))..))..))..))))..)...))))).....)))))......))))))))).))).)). ( -40.80) >DroEre_CAF1 14488 118 - 1 AAAUGUCAGGGCGCUUGUCGCCGUUGUUGCCAUAGUGGCCAUGGCCCAUGGCCCAUG--GCCCUUGUUGUUCUUGCUGUUUUUGUGGGCUCUGCAAUUUAUAAGCGACAGCCGCUGUCAG ...((.(((((((.....))))((((((((....(.((((((((((...)).)))))--))))((((.(..((..(.......)..))..).)))).......))))))))..))).)). ( -44.30) >DroYak_CAF1 15696 120 - 1 AAAUGUCAGGGCGCUUGUCGCCGUUGUUGCCAUAGUGGCCAUGGCCCAUGGCCCUUGUUGUUCUUGCUGUUUUUGCUGUUUUUGUGGGCACUGCAAUUUAUAAGCGACAGCCGCUGUCAG ...((.(((.(.(((.(((((....(((((...((((.((((((((...))))............((.......)).......)))).)))))))))......))))))))).))).)). ( -40.50) >consensus AAAUGUCAGGGCGCUUGUCGCCGUUGUUGCCACAGUGGCCAUGGCCCAUGGCCCAUG__GCCCUUGUUGUUCUUGCUGUUUUUGUGGAAUCUGCAAUUUAUAAGCGACAGCCGCUGUCAG ...((.(((.(.(((.(((((.((....((.((((.((((((((.(....).)))))..))).)))).))....))(((..(((..(...)..)))...))).))))))))).))).)). (-32.44 = -32.80 + 0.36)


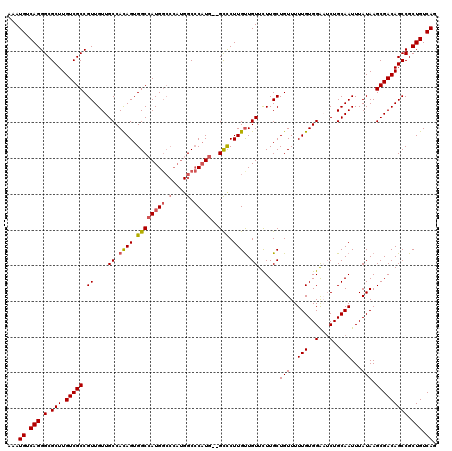
| Location | 21,043,347 – 21,043,458 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.60 |
| Mean single sequence MFE | -35.36 |
| Consensus MFE | -27.68 |
| Energy contribution | -29.12 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.547195 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21043347 111 + 22224390 AACAGCAAGAACAACAAGGAC--CAUGCGC-------CAUGGCCACUGUGGCAACAACGGCGACAAGCGCCCUGACAUUUAACAUCCGGUUGAAAUUGUUCAACGCAUUUCCGGUGCUCU ........((((((....(((--(....((-------(((((...)))))))......((((.....))))................))))....))))))...((((.....))))... ( -28.50) >DroSec_CAF1 13776 118 + 1 AACAGCAAGAACAACAAGGGC--CAUGGGCCAUGGGCCAUGGCCACUGUGGCAACAACGGCGACAAGCGCCCUGACAUUUAACAUCCGGUUGAAAUUGUUCAACGCAUUUCCGGUGCUCU ........((((((((..(((--(((((.(....).))))))))..))((....))..((((.....))))......((((((.....)))))).))))))...((((.....))))... ( -39.50) >DroSim_CAF1 14294 118 + 1 AACAGCAAGAACAACAAGGGC--CAUGGGCCAUGGGCCAUUGCCACUGUGGCAACAACGGCGACAAGCGCCCUGACAUUUAACAUCCGGUUGAAAUUGUUCAACGCAUUUCCCGUGCUCU ........((((((..(((((--(.((((((...)))).(((((....((....))..))))))).).)))))....((((((.....)))))).))))))...((((.....))))... ( -36.50) >DroEre_CAF1 14528 118 + 1 AACAGCAAGAACAACAAGGGC--CAUGGGCCAUGGGCCAUGGCCACUAUGGCAACAACGGCGACAAGCGCCCUGACAUUUAACAUCCGGUUGAAAUUGUUCAACGCAUUUCCGGUGCUCU ........((((((..(((((--(((((.(....).)))))))).)).((....))..((((.....))))......((((((.....)))))).))))))...((((.....))))... ( -39.40) >DroYak_CAF1 15736 120 + 1 AACAGCAAAAACAGCAAGAACAACAAGGGCCAUGGGCCAUGGCCACUAUGGCAACAACGGCGACAAGCGCCCUGACAUUUAACAUCCGGUUGAAAUUGUUCAACGCAUUUCCGGUGCUCU ....((.......))..((((((..(((((.....(((((((...))))))).......((.....)))))))....((((((.....)))))).))))))...((((.....))))... ( -32.90) >consensus AACAGCAAGAACAACAAGGGC__CAUGGGCCAUGGGCCAUGGCCACUGUGGCAACAACGGCGACAAGCGCCCUGACAUUUAACAUCCGGUUGAAAUUGUUCAACGCAUUUCCGGUGCUCU ........((((((..(((((..(((((.(....).)))))(((....((....))..))).......)))))....((((((.....)))))).))))))...((((.....))))... (-27.68 = -29.12 + 1.44)

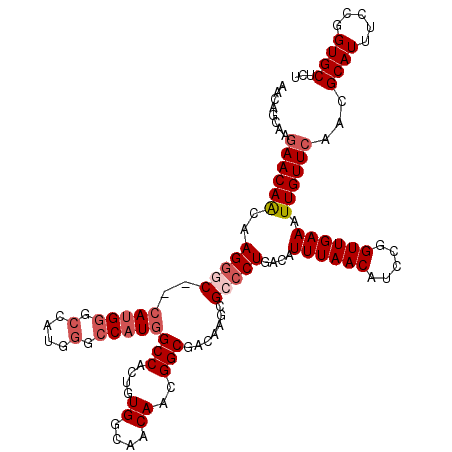
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:05:07 2006