
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 21,041,372 – 21,041,463 |
| Length | 91 |
| Max. P | 0.884948 |

| Location | 21,041,372 – 21,041,463 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.61 |
| Mean single sequence MFE | -30.97 |
| Consensus MFE | -11.40 |
| Energy contribution | -11.80 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.37 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.884948 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21041372 91 + 22224390 AGAGCUA----------------AAGGGGACGG---CGGGCAAAAUGAAAACAUUUACAGGGCAGAGUACCCACUGACCUGUCCC----------CACUCUAUGGUCCAAAGAAAAUAUG ((((...----------------..((((((((---(((...(((((....)))))...(((.......))).))).)).)))))----------).))))................... ( -24.20) >DroSec_CAF1 11799 90 + 1 AGAGCUA----------------AAGGGGUCGG---CGGGG-AAAUGAAAACAUUUACAGGGCAGAGUACCCACUGACCUGUCCC----------CUCUCUAUGGUCCAAAGAAAAUAUG .(..(((----------------.(((((....---.((((-(((((....)))))(((((.(((........))).))))))))----------)))))).)))..)............ ( -24.40) >DroSim_CAF1 12318 90 + 1 AGAGCUA----------------AAGGGGUCGG---CGGGG-AAAUGAAAACAUUUACAGGGCAGAGUACCCACUGACCUGUCCC----------CUCUCUAUGGUCCAAAGAAAAUAUG .(..(((----------------.(((((....---.((((-(((((....)))))(((((.(((........))).))))))))----------)))))).)))..)............ ( -24.40) >DroEre_CAF1 12472 119 + 1 AGAGCUGCAGAGCUACAGAGCUAAGGGGGACGGCGGUGGGG-AAAUAAAAACAUUUACAAGGCAGUGUUCCCACUGACCUGUCCCCCCUGCCCCCCUCCUUAUGCUCCAAAGAAAAUAUG ..((((....))))...((((((((((((..(((((.((((-(................((((((((....))))).))).))))).)))))..)))))))).))))............. ( -48.73) >DroYak_CAF1 13609 99 + 1 AGGGCUGAA-AGGA---------ACGGGGGGGUCGACGGGA-AAAUGAAAACAUUUACAAGGCAGAGUUCCCACUGACCUGUCCC----------CUCUUUAUGGUCCAAAGAAAAUAUG .((((((.(-((((---------..((((((((((..((((-(..((....(........).))...)))))..))))))..)))----------)))))).))))))............ ( -33.10) >consensus AGAGCUA________________AAGGGGACGG___CGGGG_AAAUGAAAACAUUUACAGGGCAGAGUACCCACUGACCUGUCCC__________CUCUCUAUGGUCCAAAGAAAAUAUG ..........................((..((.....(((..(((((....)))))...((((((.((........)))))))).............)))..))..))............ (-11.40 = -11.80 + 0.40)

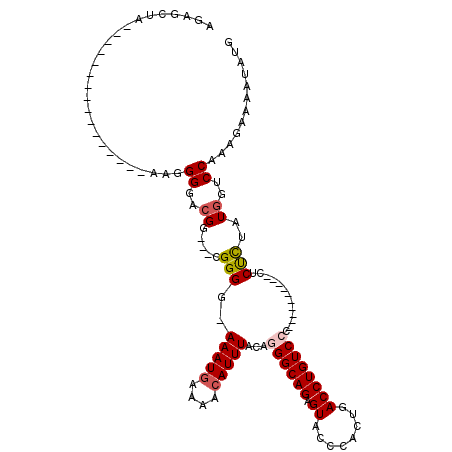
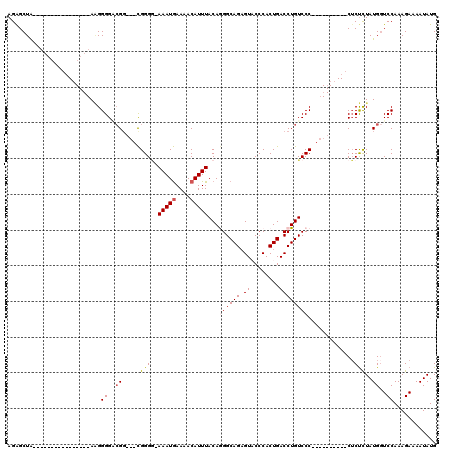
| Location | 21,041,372 – 21,041,463 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.61 |
| Mean single sequence MFE | -30.36 |
| Consensus MFE | -19.00 |
| Energy contribution | -18.40 |
| Covariance contribution | -0.60 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.855278 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21041372 91 - 22224390 CAUAUUUUCUUUGGACCAUAGAGUG----------GGGACAGGUCAGUGGGUACUCUGCCCUGUAAAUGUUUUCAUUUUGCCCG---CCGUCCCCUU----------------UAGCUCU ...................((((((----------(((((.(((..((((((.....))))...(((((....))))).))..)---))))))))..----------------..))))) ( -25.90) >DroSec_CAF1 11799 90 - 1 CAUAUUUUCUUUGGACCAUAGAGAG----------GGGACAGGUCAGUGGGUACUCUGCCCUGUAAAUGUUUUCAUUU-CCCCG---CCGACCCCUU----------------UAGCUCU .......((((((.....))))))(----------((((((((.(((.(....).))).)))))(((((....)))))-))))(---(..(.....)----------------..))... ( -23.90) >DroSim_CAF1 12318 90 - 1 CAUAUUUUCUUUGGACCAUAGAGAG----------GGGACAGGUCAGUGGGUACUCUGCCCUGUAAAUGUUUUCAUUU-CCCCG---CCGACCCCUU----------------UAGCUCU .......((((((.....))))))(----------((((((((.(((.(....).))).)))))(((((....)))))-))))(---(..(.....)----------------..))... ( -23.90) >DroEre_CAF1 12472 119 - 1 CAUAUUUUCUUUGGAGCAUAAGGAGGGGGGCAGGGGGGACAGGUCAGUGGGAACACUGCCUUGUAAAUGUUUUUAUUU-CCCCACCGCCGUCCCCCUUAGCUCUGUAGCUCUGCAGCUCU ............(((((.(((((.(((((((.(((((((((((.(((((....))))).)))))(((((....)))))-)))).))))).))))))))))))))..((((....)))).. ( -51.90) >DroYak_CAF1 13609 99 - 1 CAUAUUUUCUUUGGACCAUAAAGAG----------GGGACAGGUCAGUGGGAACUCUGCCUUGUAAAUGUUUUCAUUU-UCCCGUCGACCCCCCCGU---------UCCU-UUCAGCCCU .......((((((.....))))))(----------(((...((((..(((((((........))(((((....)))))-)))))..)))).))))..---------....-......... ( -26.20) >consensus CAUAUUUUCUUUGGACCAUAGAGAG__________GGGACAGGUCAGUGGGUACUCUGCCCUGUAAAUGUUUUCAUUU_CCCCG___CCGACCCCUU________________UAGCUCU ......(((((((.....)))))))..........((((((((.(((.(....).))).)))))(((((....)))))..)))..................................... (-19.00 = -18.40 + -0.60)

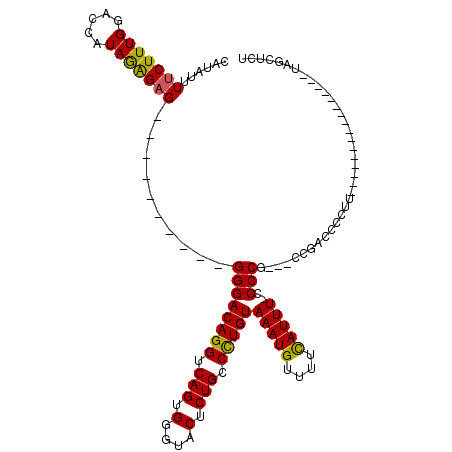
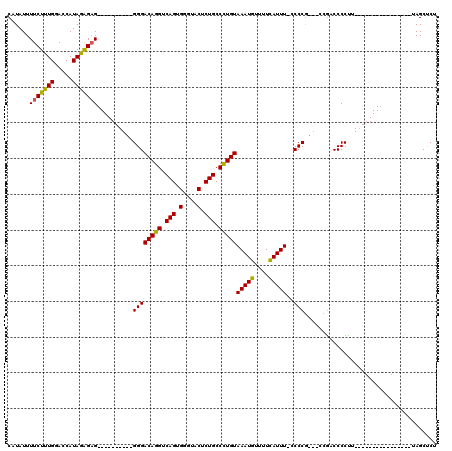
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:04:59 2006