
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 21,040,326 – 21,040,478 |
| Length | 152 |
| Max. P | 0.896199 |

| Location | 21,040,326 – 21,040,438 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.70 |
| Mean single sequence MFE | -25.48 |
| Consensus MFE | -23.40 |
| Energy contribution | -23.80 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.808881 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
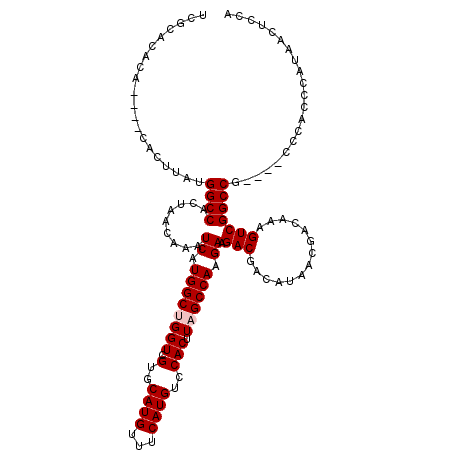
>X_DroMel_CAF1 21040326 112 - 22224390 UCGCACACA----CACUUAUGGCCACUAACAAAUCAUGGCUGGUCGUGCAUGUUUCAUGUCCACUUAGCCAAGAAGACGACAUAACGACAAAGUCGGCCG----CCCACCCAUAACUCCA .........----.......((((.........((.((((((((.(..((((...))))..))).)))))).)).(((..............))))))).----................ ( -25.04) >DroSec_CAF1 10784 112 - 1 UCGCACACA----CACUUAUGGCCACUAACAAAUCAUGGCCGGUCGUGCAUGUUUCAUGUCCACUUAGCCAAGAAGACGACAUAACGACAAAGUCGGCCG----CCCACCCAUAACUCCA ..((.....----.......(((((...........)))))(((((....((((..(((((..(((.......)))..)))))...))))....))))))----)............... ( -23.80) >DroSim_CAF1 11274 112 - 1 UCGCACACA----CACUUAUGGCCACUAACAAAUCAUGGCCGGUCGUGCAUGUUUCAUGUCCACUUAGCCAAGAAGACGACAUAACGACAAAGUCGGCCG----CCCACCCAUAACUCCA ..((.....----.......(((((...........)))))(((((....((((..(((((..(((.......)))..)))))...))))....))))))----)............... ( -23.80) >DroEre_CAF1 11481 116 - 1 UCGCACACACUCGCACUUAUGGCCACUAACAAAUCAUGGCUGGUCGUGCAUGUUUCAUGUCCACUUAGCCAAGAAGACGACAUAACGACAAAGUCGGCCG----CCCACCCAUAACUCCA ..((........))......((((.........((.((((((((.(..((((...))))..))).)))))).)).(((..............))))))).----................ ( -26.34) >DroYak_CAF1 12517 120 - 1 UCGCGCACACUCGCACUUAUGGCCACUAACAAAUCAUGGCUGGUCGUGCAUGUUUCAUGUCCACUUAGCCAAGAAGACGACAUAACGACAAAGUCGGCCGCCCGCCCACCCAUAACUCCA ..((((......))......((((.........((.((((((((.(..((((...))))..))).)))))).)).(((..............)))))))))................... ( -28.44) >consensus UCGCACACA____CACUUAUGGCCACUAACAAAUCAUGGCUGGUCGUGCAUGUUUCAUGUCCACUUAGCCAAGAAGACGACAUAACGACAAAGUCGGCCG____CCCACCCAUAACUCCA ....................((((.........((.((((((((.(..((((...))))..))).)))))).)).(((..............)))))))..................... (-23.40 = -23.80 + 0.40)

| Location | 21,040,362 – 21,040,478 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.77 |
| Mean single sequence MFE | -29.18 |
| Consensus MFE | -24.87 |
| Energy contribution | -25.67 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.896199 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21040362 116 - 22224390 AAAAAAAAUGAGAAAAACAAUAUAUGGCGGGCCGUAAGAGUCGCACACA----CACUUAUGGCCACUAACAAAUCAUGGCUGGUCGUGCAUGUUUCAUGUCCACUUAGCCAAGAAGACGA ...........................(((((((((((.((.......)----).))))))))).........((.((((((((.(..((((...))))..))).)))))).))...)). ( -29.80) >DroSec_CAF1 10820 115 - 1 AAA-AAAAUGAGAAAAACAAUAUAUGGCGGGCCGUAAGUGUCGCACACA----CACUUAUGGCCACUAACAAAUCAUGGCCGGUCGUGCAUGUUUCAUGUCCACUUAGCCAAGAAGACGA ...-...((((((....((.....((((.((((((((((((.......)----))))))))))).....((.....))))))....))....))))))(((..(((....)))..))).. ( -32.10) >DroSim_CAF1 11310 116 - 1 AAAAAAAAUGAGAAAAACAAUAUAUGGCGGGCCGUAAGAGUCGCACACA----CACUUAUGGCCACUAACAAAUCAUGGCCGGUCGUGCAUGUUUCAUGUCCACUUAGCCAAGAAGACGA .......((((((....((.....((((.(((((((((.((.......)----).))))))))).....((.....))))))....))....))))))(((..(((....)))..))).. ( -27.40) >DroEre_CAF1 11517 118 - 1 AAA--AAAUGGCAAAAACAAUAUAUGGCGGGCCGAAAGAGUCGCACACACUCGCACUUAUGGCCACUAACAAAUCAUGGCUGGUCGUGCAUGUUUCAUGUCCACUUAGCCAAGAAGACGA ...--...((((.....((.....))..((((.((((((((.......))))((((...((((((...........))))))...))))...))))..)))).....))))......... ( -27.50) >DroYak_CAF1 12557 119 - 1 AAA-AAAAUGAGAAAAACAAUAUAUGGCGGGCCGAAAGAGUCGCGCACACUCGCACUUAUGGCCACUAACAAAUCAUGGCUGGUCGUGCAUGUUUCAUGUCCACUUAGCCAAGAAGACGA ...-...((((((...........(((((((((....).))).))).))...((((...((((((...........))))))...))))...))))))(((..(((....)))..))).. ( -29.10) >consensus AAA_AAAAUGAGAAAAACAAUAUAUGGCGGGCCGUAAGAGUCGCACACA____CACUUAUGGCCACUAACAAAUCAUGGCUGGUCGUGCAUGUUUCAUGUCCACUUAGCCAAGAAGACGA ...........................(((((((((((.................))))))))).........((.((((((((.(..((((...))))..))).)))))).))...)). (-24.87 = -25.67 + 0.80)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:04:58 2006