
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 21,039,479 – 21,039,663 |
| Length | 184 |
| Max. P | 0.999904 |

| Location | 21,039,479 – 21,039,594 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.19 |
| Mean single sequence MFE | -30.49 |
| Consensus MFE | -25.04 |
| Energy contribution | -24.96 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.82 |
| SVM decision value | 2.06 |
| SVM RNA-class probability | 0.987024 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21039479 115 - 22224390 AAUUAAGCAAAUGGCUUUUUUGUUACAUUUUUUAUGGGUCCUCAAAAGUUCCCUCGGAUUUAUUAACGUAACAGAGAAGUGCUCAAUUAUGACC-----AAGUCCGAGCAAACGGAAUUU ((((.(((.....(((((((((((((.((....((((((((..............)))))))).)).)))))))))))))))).))))......-----...((((......)))).... ( -25.94) >DroSec_CAF1 10001 113 - 1 AAUUAAGGAAAUGGCUUUUUUGUUACAUUUUUGAUGGGUCCUCA--AGUUCCCUCGGACUUAUUAACGUAACAGAGAAGUGCUCAAUUGUGACC-----AAGUCCGUGCAAACGGAAUUU ......((.....(((((((((((((....(((((((((((...--.........))))))))))).)))))))))))))((......))..))-----...(((((....))))).... ( -33.40) >DroSim_CAF1 10495 113 - 1 AAUUAAGGAAAUGGCUUUUUUGUUACAUUUUUGAUGGGUCCUCA--AGUUGCCUUGGAUUUAUUAACGUAACAGAGAAGUGCUCAAUUAUGACC-----AAGUCCGUGCAAACGGAAUUU ......((.....(((((((((((((....(((((((((((...--.........))))))))))).)))))))))))))..(((....)))))-----...(((((....))))).... ( -31.40) >DroEre_CAF1 10730 113 - 1 AAUUAGGGAAAUGGCUUUUUUGUUACAUUUUUUAUGAGCCCUCA--AAUUCCCUCGGAUUUAUUAAUGUAACAGAGAAGUGCUCAAUUAUGGCC-----GAGUCCGUGCAAACGGAAUUU .....(((.....((((((((((((((((....((((..((...--.........))..)))).)))))))))))))))).)))..........-----...(((((....))))).... ( -28.30) >DroYak_CAF1 11687 118 - 1 AAUUAGGGAAAUGGCUUUUUUGUUACAUUUUUUAUGGGUCUUCA--AAUCCCCUCGGAUUUAUUAAUGUAACAGAGAAGUGCUCAAUUAUGGCCAAGCCAAGUCCGUGCAAACGGAAUUU .....(((.....((((((((((((((((....((((((((...--.........)))))))).)))))))))))))))).))).....((((...))))..(((((....))))).... ( -33.40) >consensus AAUUAAGGAAAUGGCUUUUUUGUUACAUUUUUUAUGGGUCCUCA__AGUUCCCUCGGAUUUAUUAACGUAACAGAGAAGUGCUCAAUUAUGACC_____AAGUCCGUGCAAACGGAAUUU .............(((((((((((((.((....((((((((..............)))))))).)).)))))))))))))......................(((((....))))).... (-25.04 = -24.96 + -0.08)



| Location | 21,039,514 – 21,039,629 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.56 |
| Mean single sequence MFE | -26.04 |
| Consensus MFE | -18.41 |
| Energy contribution | -17.69 |
| Covariance contribution | -0.72 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.817680 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21039514 115 + 22224390 ACUUCUCUGUUACGUUAAUAAAUCCGAGGGAACUUUUGAGGACCCAUAAAAAAUGUAACAAAAAAGCCAUUUGCUUAAUUUGCAAAUAGUUUCUUCAAGCACGAAGCUGAAAGGG----- .......(((((((((...........(((..((....))..)))......)))))))))...((((.((((((.......)))))).))))(((..(((.....)))..)))..----- ( -22.43) >DroSec_CAF1 10036 113 + 1 ACUUCUCUGUUACGUUAAUAAGUCCGAGGGAACU--UGAGGACCCAUCAAAAAUGUAACAAAAAAGCCAUUUCCUUAAUUUGGAAAUAGUUUCUUCAAGCAAGAAGCUGAAAAGG----- .(((.(((((((((((.....(((((((....))--)..))))........)))))))))........((((((.......))))))((((((((.....)))))))))).))).----- ( -27.42) >DroSim_CAF1 10530 113 + 1 ACUUCUCUGUUACGUUAAUAAAUCCAAGGCAACU--UGAGGACCCAUCAAAAAUGUAACAAAAAAGCCAUUUCCUUAAUUUGGAAAUAGUUCCUUCAAGCAAGAAGCUGAAAAGG----- .......(((((((((........((((....))--)).(....)......)))))))))....(((.((((((.......)))))).)))((((..(((.....)))...))))----- ( -25.50) >DroEre_CAF1 10765 113 + 1 ACUUCUCUGUUACAUUAAUAAAUCCGAGGGAAUU--UGAGGGCUCAUAAAAAAUGUAACAAAAAAGCCAUUUCCCUAAUUUGGAAAUAGUUUCCCCAAGCGUGAAGCCGAGAAGG----- .((((((.(((.(((.......((((((((((..--....((((....................))))..))))))....))))....((((....))))))).))).)))))).----- ( -26.15) >DroYak_CAF1 11727 118 + 1 ACUUCUCUGUUACAUUAAUAAAUCCGAGGGGAUU--UGAAGACCCAUAAAAAAUGUAACAAAAAAGCCAUUUCCCUAAUUUGGAAAUAGUUUCUCCAAGCAUGAAGCUUGGAAUAUUUAG .......(((((((((...((((((....)))))--)(......)......)))))))))...((((.((((((.......)))))).)))).(((((((.....)))))))........ ( -28.70) >consensus ACUUCUCUGUUACGUUAAUAAAUCCGAGGGAACU__UGAGGACCCAUAAAAAAUGUAACAAAAAAGCCAUUUCCUUAAUUUGGAAAUAGUUUCUUCAAGCAAGAAGCUGAAAAGG_____ .((((..(((((((((...........(((..((....))..)))......)))))))))...((((.((((((.......)))))).))))..........)))).............. (-18.41 = -17.69 + -0.72)



| Location | 21,039,514 – 21,039,629 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.56 |
| Mean single sequence MFE | -35.23 |
| Consensus MFE | -31.52 |
| Energy contribution | -29.80 |
| Covariance contribution | -1.72 |
| Combinations/Pair | 1.24 |
| Mean z-score | -4.39 |
| Structure conservation index | 0.89 |
| SVM decision value | 4.47 |
| SVM RNA-class probability | 0.999904 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21039514 115 - 22224390 -----CCCUUUCAGCUUCGUGCUUGAAGAAACUAUUUGCAAAUUAAGCAAAUGGCUUUUUUGUUACAUUUUUUAUGGGUCCUCAAAAGUUCCCUCGGAUUUAUUAACGUAACAGAGAAGU -----..((((.(((.....))).))))...((((((((.......))))))))((((((((((((.((....((((((((..............)))))))).)).)))))))))))). ( -31.44) >DroSec_CAF1 10036 113 - 1 -----CCUUUUCAGCUUCUUGCUUGAAGAAACUAUUUCCAAAUUAAGGAAAUGGCUUUUUUGUUACAUUUUUGAUGGGUCCUCA--AGUUCCCUCGGACUUAUUAACGUAACAGAGAAGU -----..((((((((.....)).))))))..((((((((.......))))))))((((((((((((....(((((((((((...--.........))))))))))).)))))))))))). ( -36.80) >DroSim_CAF1 10530 113 - 1 -----CCUUUUCAGCUUCUUGCUUGAAGGAACUAUUUCCAAAUUAAGGAAAUGGCUUUUUUGUUACAUUUUUGAUGGGUCCUCA--AGUUGCCUUGGAUUUAUUAACGUAACAGAGAAGU -----(((((..(((.....))).)))))..((((((((.......))))))))((((((((((((....(((((((((((...--.........))))))))))).)))))))))))). ( -36.60) >DroEre_CAF1 10765 113 - 1 -----CCUUCUCGGCUUCACGCUUGGGGAAACUAUUUCCAAAUUAGGGAAAUGGCUUUUUUGUUACAUUUUUUAUGAGCCCUCA--AAUUCCCUCGGAUUUAUUAAUGUAACAGAGAAGU -----..((((((((.....)).))))))..((((((((.......))))))))(((((((((((((((....((((..((...--.........))..)))).))))))))))))))). ( -31.90) >DroYak_CAF1 11727 118 - 1 CUAAAUAUUCCAAGCUUCAUGCUUGGAGAAACUAUUUCCAAAUUAGGGAAAUGGCUUUUUUGUUACAUUUUUUAUGGGUCUUCA--AAUCCCCUCGGAUUUAUUAAUGUAACAGAGAAGU .......((((((((.....))))))))...((((((((.......))))))))(((((((((((((((....((((((((...--.........)))))))).))))))))))))))). ( -39.40) >consensus _____CCUUUUCAGCUUCAUGCUUGAAGAAACUAUUUCCAAAUUAAGGAAAUGGCUUUUUUGUUACAUUUUUUAUGGGUCCUCA__AGUUCCCUCGGAUUUAUUAACGUAACAGAGAAGU .......((((.(((.....))).))))...((((((((.......))))))))((((((((((((.((....((((((((..............)))))))).)).)))))))))))). (-31.52 = -29.80 + -1.72)

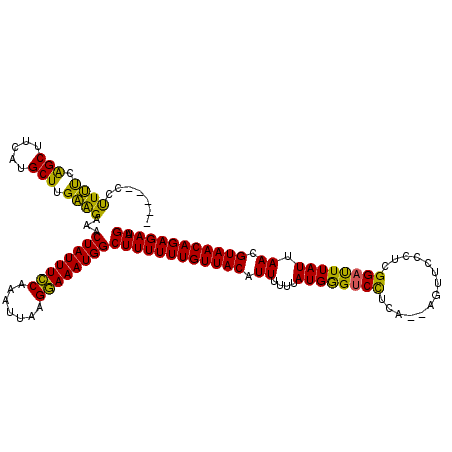
| Location | 21,039,554 – 21,039,663 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.55 |
| Mean single sequence MFE | -23.18 |
| Consensus MFE | -15.96 |
| Energy contribution | -15.96 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.943085 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21039554 109 - 22224390 UAUUGGAUUGUUGUGCUUAAUGUAGCUUC---CUUUC--------CCCUUUCAGCUUCGUGCUUGAAGAAACUAUUUGCAAAUUAAGCAAAUGGCUUUUUUGUUACAUUUUUUAUGGGUC ..........(..((...((((((((...---.....--------..((((.(((.....))).))))...((((((((.......)))))))).......))))))))...))..)... ( -22.50) >DroSec_CAF1 10074 109 - 1 UAUUGGAUUGUUCUGCUUAAUGUAGCUUG---CUUUC--------CCUUUUCAGCUUCUUGCUUGAAGAAACUAUUUCCAAAUUAAGGAAAUGGCUUUUUUGUUACAUUUUUGAUGGGUC ....(((..((.((((.....))))...)---)..))--------)((..(((((.....))..((((((.((((((((.......)))))))).))))))..........)))..)).. ( -23.70) >DroSim_CAF1 10568 109 - 1 UAUUGGAUUGUUCUGCUUAAUGUAGCUUG---CUUUC--------CCUUUUCAGCUUCUUGCUUGAAGGAACUAUUUCCAAAUUAAGGAAAUGGCUUUUUUGUUACAUUUUUGAUGGGUC .....((((..((.....((((((((...---.....--------(((((..(((.....))).)))))..((((((((.......)))))))).......))))))))...))..)))) ( -25.30) >DroEre_CAF1 10803 91 - 1 ------------------AAUUUCUAUUG---CUUUG--------CCUUCUCGGCUUCACGCUUGGGGAAACUAUUUCCAAAUUAGGGAAAUGGCUUUUUUGUUACAUUUUUUAUGAGCC ------------------..(((((...(---(...(--------((.....))).....))...))))).((((((((.......)))))))).......(((.(((.....)))))). ( -17.80) >DroYak_CAF1 11765 102 - 1 ------------------GAUUUCUAUCGGAACUGUGCUGCUAAAUAUUCCAAGCUUCAUGCUUGGAGAAACUAUUUCCAAAUUAGGGAAAUGGCUUUUUUGUUACAUUUUUUAUGGGUC ------------------....(((((..(((.((((..........((((((((.....))))))))...((((((((.......)))))))).........)))).)))..))))).. ( -26.60) >consensus UAUUGGAUUGUU_UGCUUAAUGUAGCUUG___CUUUC________CCUUUUCAGCUUCAUGCUUGAAGAAACUAUUUCCAAAUUAAGGAAAUGGCUUUUUUGUUACAUUUUUUAUGGGUC ..................((((((((.....................((((.(((.....))).))))...((((((((.......)))))))).......))))))))........... (-15.96 = -15.96 + -0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:04:56 2006