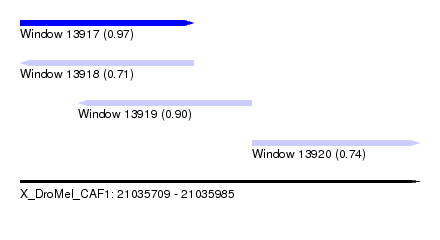
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 21,035,709 – 21,035,985 |
| Length | 276 |
| Max. P | 0.973978 |

| Location | 21,035,709 – 21,035,829 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.25 |
| Mean single sequence MFE | -30.62 |
| Consensus MFE | -27.32 |
| Energy contribution | -27.36 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.973978 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21035709 120 + 22224390 GCUCAACAUUUCUUCGUUAUUAAAUGGAUUUCUUCGCUGACUCGAAAUUGUAUGCAUACGUGCUCCAUUUGCACAUAUUUCUUAAUGGGUCAGCGCCCCGCGGAGUCCACUCCCCGCCCA ((..(((........)))......((((((((...(((((((((.....(((....)))((((.......))))...........)))))))))((...))))))))))......))... ( -31.00) >DroSec_CAF1 6341 120 + 1 GCUCAACAUUUCUUCGUUAUUAAAUGGAUUUCUUCGCUGACUCGAAAUUGUAUGCAUACGUGCUCCAUUUGCACAUAUUUCUUAAUGGGUCAGCGCCCCGCGGAGUCCACUCCCCGCCCA ((..(((........)))......((((((((...(((((((((.....(((....)))((((.......))))...........)))))))))((...))))))))))......))... ( -31.00) >DroSim_CAF1 6878 120 + 1 GCUCAACAUUUCUUCGUUAUUAAAUGGAUUUCUUCGCUGACUCGAAAUUGUAUGCAUACGUGCUCCAUUUGCACAUAUUUCUUAAUGGGUCAGCGCCCCGCGGAGUCCACUCCCCGCCCA ((..(((........)))......((((((((...(((((((((.....(((....)))((((.......))))...........)))))))))((...))))))))))......))... ( -31.00) >DroEre_CAF1 6581 120 + 1 GCUUAGCUGUUCCUCGUUUCUAAAUGGAUUUCCUUGCUGACUCGGAAUUGUAUGCAUAUGUGCUCCAUUUGCACAUUUUUCUUAAUGGGUCAGCGCCCCGCGGAGUCCACUCCCCGCCCA ((...((...(((............))).......((((((((((((..((.((((.(((.....))).)))).))..))))....))))))))))...))((((....))))....... ( -32.30) >DroYak_CAF1 7030 120 + 1 GCUUGACUGUUCGUCGUUUUUAAAUGGAUUUCUUCAGUGACUCGAAAUUGUAUGCAUAUGUGCUCCAUUUGCACAUAUUUCUUAAUGGGUCAGCGCCCCGCAGAGUCCACACCCCGCCCA ((..(((.....))).........(((((((......(((((((....((....))(((((((.......)))))))........)))))))(((...))).)))))))......))... ( -27.80) >consensus GCUCAACAUUUCUUCGUUAUUAAAUGGAUUUCUUCGCUGACUCGAAAUUGUAUGCAUACGUGCUCCAUUUGCACAUAUUUCUUAAUGGGUCAGCGCCCCGCGGAGUCCACUCCCCGCCCA ((..(((........)))......((((((((...(((((((((.....(((....)))((((.......))))...........)))))))))((...))))))))))......))... (-27.32 = -27.36 + 0.04)

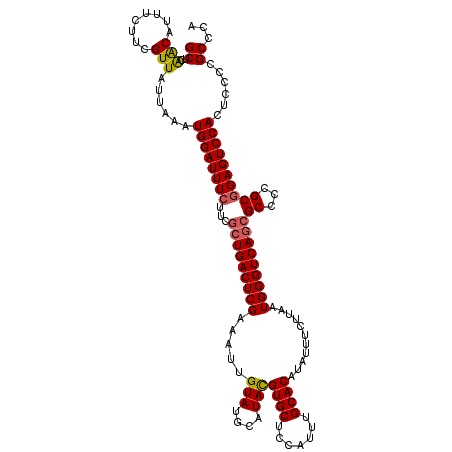

| Location | 21,035,709 – 21,035,829 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.25 |
| Mean single sequence MFE | -33.74 |
| Consensus MFE | -30.34 |
| Energy contribution | -30.58 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.711262 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21035709 120 - 22224390 UGGGCGGGGAGUGGACUCCGCGGGGCGCUGACCCAUUAAGAAAUAUGUGCAAAUGGAGCACGUAUGCAUACAAUUUCGAGUCAGCGAAGAAAUCCAUUUAAUAACGAAGAAAUGUUGAGC ...((((((......))))))(((.(((((((.......(((((.((((((.(((.....))).))))))..)))))..)))))))......)))......(((((......)))))... ( -34.60) >DroSec_CAF1 6341 120 - 1 UGGGCGGGGAGUGGACUCCGCGGGGCGCUGACCCAUUAAGAAAUAUGUGCAAAUGGAGCACGUAUGCAUACAAUUUCGAGUCAGCGAAGAAAUCCAUUUAAUAACGAAGAAAUGUUGAGC ...((((((......))))))(((.(((((((.......(((((.((((((.(((.....))).))))))..)))))..)))))))......)))......(((((......)))))... ( -34.60) >DroSim_CAF1 6878 120 - 1 UGGGCGGGGAGUGGACUCCGCGGGGCGCUGACCCAUUAAGAAAUAUGUGCAAAUGGAGCACGUAUGCAUACAAUUUCGAGUCAGCGAAGAAAUCCAUUUAAUAACGAAGAAAUGUUGAGC ...((((((......))))))(((.(((((((.......(((((.((((((.(((.....))).))))))..)))))..)))))))......)))......(((((......)))))... ( -34.60) >DroEre_CAF1 6581 120 - 1 UGGGCGGGGAGUGGACUCCGCGGGGCGCUGACCCAUUAAGAAAAAUGUGCAAAUGGAGCACAUAUGCAUACAAUUCCGAGUCAGCAAGGAAAUCCAUUUAGAAACGAGGAACAGCUAAGC ...((((((......))))))..(((((((((.(.....(((...((((((.(((.....))).))))))...))).).))))))..((....))..................))).... ( -33.30) >DroYak_CAF1 7030 120 - 1 UGGGCGGGGUGUGGACUCUGCGGGGCGCUGACCCAUUAAGAAAUAUGUGCAAAUGGAGCACAUAUGCAUACAAUUUCGAGUCACUGAAGAAAUCCAUUUAAAAACGACGAACAGUCAAGC ((((((((((....)))))))(((.......))).....(((((.((((((.(((.....))).))))))..)))))................))).........(((.....))).... ( -31.60) >consensus UGGGCGGGGAGUGGACUCCGCGGGGCGCUGACCCAUUAAGAAAUAUGUGCAAAUGGAGCACGUAUGCAUACAAUUUCGAGUCAGCGAAGAAAUCCAUUUAAUAACGAAGAAAUGUUGAGC (((((((((......))))))....(((((((.......(((((.((((((.(((.....))).))))))..)))))..))))))).......)))........................ (-30.34 = -30.58 + 0.24)


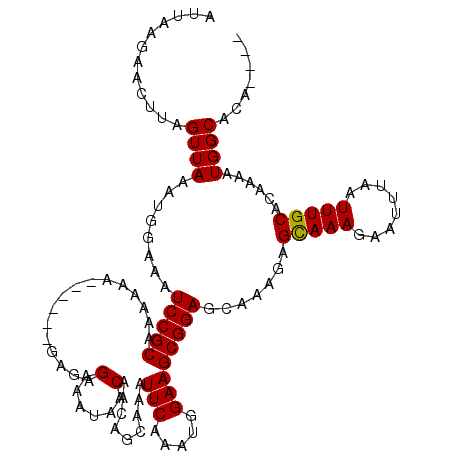
| Location | 21,035,749 – 21,035,869 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.33 |
| Mean single sequence MFE | -40.76 |
| Consensus MFE | -39.82 |
| Energy contribution | -39.82 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.898043 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21035749 120 - 22224390 CAGGCAGCCCCUUGACAACUGUCACUGGCAGGACAUUAUAUGGGCGGGGAGUGGACUCCGCGGGGCGCUGACCCAUUAAGAAAUAUGUGCAAAUGGAGCACGUAUGCAUACAAUUUCGAG ..((((((((((......((((.....))))............((((((......)))))))))).)))).))......(((((.((((((.(((.....))).))))))..)))))... ( -41.20) >DroSec_CAF1 6381 120 - 1 CAGGCAGCCCCUUGACAACUGUCACCGGCAGGACAUUAUAUGGGCGGGGAGUGGACUCCGCGGGGCGCUGACCCAUUAAGAAAUAUGUGCAAAUGGAGCACGUAUGCAUACAAUUUCGAG ..((((((((((......((((.....))))............((((((......)))))))))).)))).))......(((((.((((((.(((.....))).))))))..)))))... ( -41.20) >DroSim_CAF1 6918 120 - 1 CAGGCAGCCCCUUGACAACUGUCACCGGCAGGACAUUAUAUGGGCGGGGAGUGGACUCCGCGGGGCGCUGACCCAUUAAGAAAUAUGUGCAAAUGGAGCACGUAUGCAUACAAUUUCGAG ..((((((((((......((((.....))))............((((((......)))))))))).)))).))......(((((.((((((.(((.....))).))))))..)))))... ( -41.20) >DroEre_CAF1 6621 120 - 1 CAGGCAGCCCCUUGACAACUGUCACUGGCAGGACAUUAUAUGGGCGGGGAGUGGACUCCGCGGGGCGCUGACCCAUUAAGAAAAAUGUGCAAAUGGAGCACAUAUGCAUACAAUUCCGAG ..((((((((((......((((.....))))............((((((......)))))))))).)))).))......(((...((((((.(((.....))).))))))...))).... ( -38.90) >DroYak_CAF1 7070 120 - 1 CAGGCAGCCCCUCGACAACUGUCACUGGCAGGACAUUAUAUGGGCGGGGUGUGGACUCUGCGGGGCGCUGACCCAUUAAGAAAUAUGUGCAAAUGGAGCACAUAUGCAUACAAUUUCGAG ..((((((.((((.....((((.....))))............(((((((....))))))))))).)))).))......(((((.((((((.(((.....))).))))))..)))))... ( -41.30) >consensus CAGGCAGCCCCUUGACAACUGUCACUGGCAGGACAUUAUAUGGGCGGGGAGUGGACUCCGCGGGGCGCUGACCCAUUAAGAAAUAUGUGCAAAUGGAGCACGUAUGCAUACAAUUUCGAG ..((((((((((......((((.....))))............((((((......)))))))))).)))).))......(((((.((((((.(((.....))).))))))..)))))... (-39.82 = -39.82 + 0.00)



| Location | 21,035,869 – 21,035,985 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.23 |
| Mean single sequence MFE | -15.68 |
| Consensus MFE | -13.50 |
| Energy contribution | -13.34 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.744657 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21035869 116 + 22224390 AUUAAGAACUUAGUUAAAUGGAAAUCCGCAAAAAAAAAAAACAGGGAAAUAACAACAGCAAAUUCAAAUGGAAGCGGAGCAAAGAGUAAAGAAUUUAAUUUGCACAAAAUGGCACA---- ............((((........(((((..............(........).........(((.....))))))))(((((...(((.....))).)))))......))))...---- ( -13.90) >DroSec_CAF1 6501 110 + 1 AUUAAGAACUUAGUUAAAUAGAAAUCCGCAAAAAA------GAGAGAAAUAACAACAGCAAAUUCAAAUGGAAGCGGAGCAAAGAGCAAAGAAUUUAAUUUGCACAAAAUGGCACA---- ............((((....(...(((((......------(..(....)..).........(((.....)))))))).).....(((((........)))))......))))...---- ( -14.10) >DroSim_CAF1 7038 110 + 1 AUUAAGAACUUAGUUAAAUGGAAAUCCGCAAAAAA------GAGAGAAAUAACAACAGCAAAUUCAAAUGGAAGCGGAGCAAAGAGCAAAGAAUUUAAUUUGCACAAAAUGGCACA---- ............((((........(((((......------(..(....)..).........(((.....)))))))).......(((((........)))))......))))...---- ( -14.00) >DroEre_CAF1 6741 110 + 1 AUUAAGAACUUAGUUAAAUGGAAAUCCGCAAAAAAA------CGGGGAGUAACAACAACAAAUUCAAAUGGAAGCGGAGCAAAGAGCAAAGAAUUUAAUUUGCAGAACAUGGCACA---- ............((...(((....(((((......(------(.....))............(((.....)))))))).......(((((........)))))....)))...)).---- ( -15.30) >DroYak_CAF1 7190 117 + 1 AUUAAGAACUUAGUUAAAUGGAAAUCCGCAAAAAAUC---CGCGAGGAAUAACAACAACAAAUUCAAAUGGAAGCGGAGCAAAGAGCAAAGAAUUUAAUUUGCACAAAAUGGCACACACA ............((((........(((((......((---(....)))..............(((.....)))))))).......(((((........)))))......))))....... ( -21.10) >consensus AUUAAGAACUUAGUUAAAUGGAAAUCCGCAAAAAA______GAGAGAAAUAACAACAGCAAAUUCAAAUGGAAGCGGAGCAAAGAGCAAAGAAUUUAAUUUGCACAAAAUGGCACA____ ............((((........(((((................(......).........(((.....)))))))).......(((((........)))))......))))....... (-13.50 = -13.34 + -0.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:04:52 2006