
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 21,035,121 – 21,035,385 |
| Length | 264 |
| Max. P | 0.760168 |

| Location | 21,035,121 – 21,035,225 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 90.73 |
| Mean single sequence MFE | -25.17 |
| Consensus MFE | -19.92 |
| Energy contribution | -19.70 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.637407 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21035121 104 + 22224390 AAGAAGAAGAAGCAGCAGCGGCAGCAAAAAACUAUUCCCGCAGCUCAUUAAAAACCCAAAGGGUUAAAAUAGUGAGGGAGGGAGGCGUCACAAUGCAGUAUGCA ..............(((((....)).....(((.(((((.(..(((((((..(((((...)))))....))))))).).)))))((((....))))))).))). ( -27.50) >DroSim_CAF1 6295 94 + 1 AA------GAAGUAGCAGCGGCAGCGAAAAACUAUUCCCGCAGCUCAUUAAAAACCCAAAGGGUUAAAA-AGUGAGG---GGAGGCGUCACAAUGCAGUAUGCA ..------......(((((....)).....(((.(((((....((((((...(((((...)))))....-)))))))---))))((((....))))))).))). ( -23.60) >DroYak_CAF1 6345 94 + 1 AA------GAAGCAGCAUCGGCAGCAAAAAACUAUUCCCGCAGCUCAUUAAAAACCCAAAGGGUUAAAA-AGUGAGG---GGAGGCGCCACAAUGCAGUAUGCA ..------...(((((((.(((.((.........(((((....((((((...(((((...)))))....-)))))))---)))))))))...))))....))). ( -24.40) >consensus AA______GAAGCAGCAGCGGCAGCAAAAAACUAUUCCCGCAGCUCAUUAAAAACCCAAAGGGUUAAAA_AGUGAGG___GGAGGCGUCACAAUGCAGUAUGCA ..............(((...((.(((........((((.....((((((...(((((...))))).....))))))....)))).........))).)).))). (-19.92 = -19.70 + -0.22)

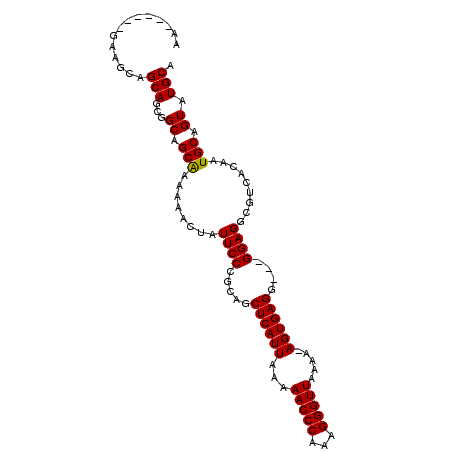

| Location | 21,035,153 – 21,035,265 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.38 |
| Mean single sequence MFE | -28.98 |
| Consensus MFE | -25.09 |
| Energy contribution | -24.87 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.760168 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21035153 112 + 22224390 UAUUCCCGCAGCUCAUUAAAAACCCAAAGGGUUAAAAUAGUGAGGGAGGGAGGCGUCAC--------AAUGCAGUAUGCAAAACAAGGCAAACAACAACAAUAAGCGUCGGAGCCGGAGA ...((((.(..(((((((..(((((...)))))....))))))).).))))(((.((.(--------.((((.((.(((........))).))...(....)..)))).))))))..... ( -28.40) >DroSim_CAF1 6321 108 + 1 UAUUCCCGCAGCUCAUUAAAAACCCAAAGGGUUAAAA-AGUGAGG---GGAGGCGUCAC--------AAUGCAGUAUGCAAAACAAGGCAAACAACAACAAUAAGCGUCGGAGCCGGAGA ..((((.((..((((((...(((((...)))))....-)))))).---.((.((.....--------..((..((.(((........))).))..)).......)).))...)).)))). ( -26.04) >DroEre_CAF1 5940 116 + 1 UAUUCCCGCAGCUCAUUAAAAACCCAAAGGGUUAAAA-AGUGAGG---GGCGGCGCCACAAUGCCACAAUGCAGUAUGCAAAACAAGGCAAACAACAACAAUAAGCGUCGGAGCUGGAGA ..((((.((..((((((...(((((...)))))....-)))))).---..((((((.....(((......)))((.(((........))).))...........))))))..)).)))). ( -32.50) >consensus UAUUCCCGCAGCUCAUUAAAAACCCAAAGGGUUAAAA_AGUGAGG___GGAGGCGUCAC________AAUGCAGUAUGCAAAACAAGGCAAACAACAACAAUAAGCGUCGGAGCCGGAGA ..((((.((..((((((...(((((...))))).....)))))).......(((((.............(((.....)))......(.....)...........)))))...)).)))). (-25.09 = -24.87 + -0.22)

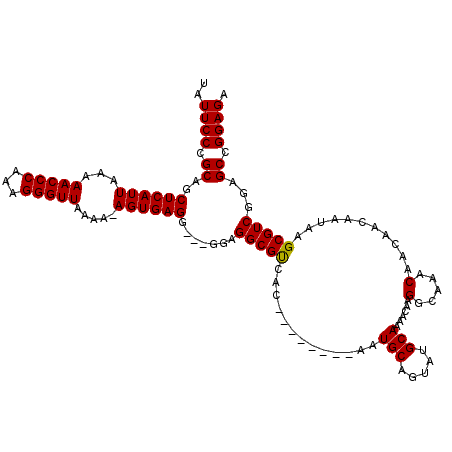

| Location | 21,035,265 – 21,035,385 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.23 |
| Mean single sequence MFE | -26.32 |
| Consensus MFE | -22.42 |
| Energy contribution | -22.50 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.718254 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21035265 120 + 22224390 AAUUGAAACUAGGCUGCCGCAAAAAAGCGACGGACAUAUUGGUGUGCGAGCGAGCAGAGAGGCUAGUGGAGAAAGUGAGCAAAGUUAAACAAUUGAAAUUGAAAUUAGUUGGUGAGCGUA ..(((..(((...((.((((......((..((.((((....)))).)).)).(((......))).))))))..)))...))).(((...(((((((........)))))))...)))... ( -27.20) >DroSec_CAF1 5880 117 + 1 AAUUGAAACUAGGCUGCCGCAAAA-AGGGACGGACAUAGGGGUGUGCGAGCGAGCAG--GGGCUAGUGGAGAAAGUGAGCAAAGUUAAACAAUUGAAAUUGAAAUUAGUUGGUGAGCGUA ..(((..(((...((.((((....-.....((.((((....)))).))(((......--..))).))))))..)))...))).(((...(((((((........)))))))...)))... ( -24.70) >DroSim_CAF1 6429 117 + 1 AAUUGAAACUAGGCUGCCGCAAAA-AGGGACGGACAUAGGGGUGUGCGAGCGAGCAG--GGGCUAGUGGAGAAAGUGAGCAAAGUUAAACAAUUGAAAUUGAAAUUAGUUGGUGAGCGUA ..(((..(((...((.((((....-.....((.((((....)))).))(((......--..))).))))))..)))...))).(((...(((((((........)))))))...)))... ( -24.70) >DroEre_CAF1 6056 116 + 1 AAUUGAAACUAGGCUGCCGCAGAA-AGGGACGGACAU-GGGGUGUGCGAGCGAGCCG--GCGCUAAUGGAGAAAGUGAGCAAAGUUAAACAAUUGAAAUUGAAAUUAGUUGGCGAGCGUA ...........((((((((((.(.-..(......)..-....).)))).)).)))).--(((((..........(....)...(((((..((((........))))..))))).))))). ( -24.30) >DroYak_CAF1 6479 117 + 1 AAUUGAAACUAGGCUGCCGCAGAA-AGGGACGGACACAGGGGUGUGCGAGCGAGGCG--GCACUAGUGGAGAAAGUGGGCAAAGUUAAAUAAUUGAAAUUGAAAUUAGUUGGCGAGCGUA ..((...(((((((((((.(....-..)..((.((((....)))).)).....))))--)).)))))...))......((...(((((.(((((........))))).)))))..))... ( -30.70) >consensus AAUUGAAACUAGGCUGCCGCAAAA_AGGGACGGACAUAGGGGUGUGCGAGCGAGCAG__GGGCUAGUGGAGAAAGUGAGCAAAGUUAAACAAUUGAAAUUGAAAUUAGUUGGUGAGCGUA ..(((..(((...((.((((..........((.((((....)))).))....(((......))).))))))..)))...))).(((...(((((((........)))))))...)))... (-22.42 = -22.50 + 0.08)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:04:49 2006