
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 21,033,818 – 21,033,938 |
| Length | 120 |
| Max. P | 0.993892 |

| Location | 21,033,818 – 21,033,938 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.33 |
| Mean single sequence MFE | -40.88 |
| Consensus MFE | -36.72 |
| Energy contribution | -36.44 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.43 |
| SVM RNA-class probability | 0.993892 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
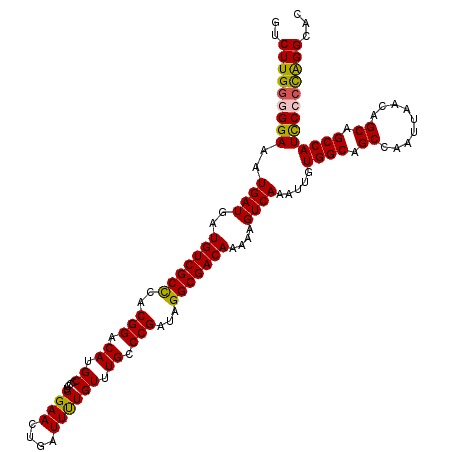
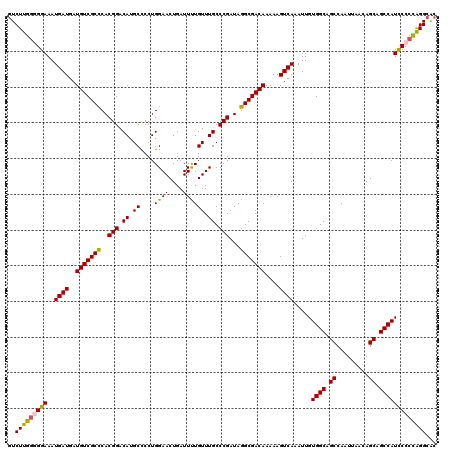
>X_DroMel_CAF1 21033818 120 + 22224390 GUCUUGGGGGAAAUGAUGAUGUCGCCCACGGACAUGCUCCUGGAACUGAUUUUGUUUGCCCGAUAGGCGACAAAAAGUCAAAUUGUGGCAGCCAAUUAACAGCAGCCAUCCCCUAGGCGC ((((.((((((..((((..(((((((..(((.((.((................)).)).)))...)))))))....)))).....((((.((.........)).)))))))))))))).. ( -41.29) >DroSec_CAF1 4468 119 + 1 GUCUUGG-GGAAAUGAUGAUGUCGCUCACGGACAUGCCCCUGGAACUGAUUUUGUUUGCCCGAUAGGCGACAAAAAGUCAAAUUGUGGCAGCCAAUUAACAGCAGCCAUCCCCCAGGCAC ((((.((-(((..((((..(((((((..(((.((......))((((.......))))..)))...)))))))....)))).....((((.((.........)).))))))))).)))).. ( -38.20) >DroSim_CAF1 5014 119 + 1 GUCUUGG-GGAAAUGAUGAUGUCGCCCACGGACAUGCCCCUGGAACUGAUUUUGUUUGCCCGAUAGGCGACAAAAAGUCAAAUUGUGGCAGCCAAUUAACAGCAGCCAUCCCCCAGGCAC ((((.((-(((..((((..(((((((..(((.((......))((((.......))))..)))...)))))))....)))).....((((.((.........)).))))))))).)))).. ( -40.10) >DroEre_CAF1 4574 119 + 1 GUCUUG-GGGAAAUGAUGAUGUCGCCCACGGACAUGCCCCUGGAACUGAUUCUGUUUGCCCGAUAGGCGACAAAAAGUCAAAUUGUGGCAGCCAAUUAACAGCAGCCAUUCCCCAGGCAC ((((.(-(((((.((((..(((((((..(((.((.((....((((....)))))).)).)))...)))))))....)))).....((((.((.........)).)))))))))))))).. ( -42.00) >DroYak_CAF1 5028 120 + 1 AUCUUGGGGGAAAUGAUGAUGUCGCCCACGGACAUGCCCCUGGAACUGAUUCUGUUUGCCCGAUAGGCGACAAAAAGUCAAAUUGUGGCAGCCAAUUAACAGCAGCCAUCCCCCGGGCAC ..(((((((((..((((..(((((((..(((.((.((....((((....)))))).)).)))...)))))))....)))).....((((.((.........)).)))))))))))))... ( -42.80) >consensus GUCUUGGGGGAAAUGAUGAUGUCGCCCACGGACAUGCCCCUGGAACUGAUUUUGUUUGCCCGAUAGGCGACAAAAAGUCAAAUUGUGGCAGCCAAUUAACAGCAGCCAUCCCCCAGGCAC ..(((((((((..((((..(((((((..(((.((.((....((((....)))))).)).)))...)))))))....)))).....((((.((.........)).)))))))))))))... (-36.72 = -36.44 + -0.28)

| Location | 21,033,818 – 21,033,938 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.33 |
| Mean single sequence MFE | -43.14 |
| Consensus MFE | -36.84 |
| Energy contribution | -38.04 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.83 |
| SVM RNA-class probability | 0.979109 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
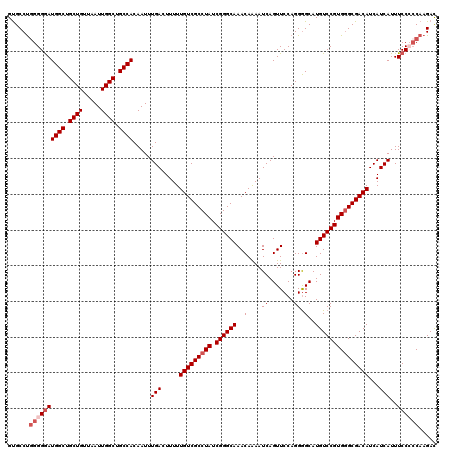
>X_DroMel_CAF1 21033818 120 - 22224390 GCGCCUAGGGGAUGGCUGCUGUUAAUUGGCUGCCACAAUUUGACUUUUUGUCGCCUAUCGGGCAAACAAAAUCAGUUCCAGGAGCAUGUCCGUGGGCGACAUCAUCAUUUCCCCCAAGAC ....((.((((.((((.((((.....)))).)))).....(((.....(((((((((.((((((..........((((...)))).)))))))))))))))...)))....)))).)).. ( -41.90) >DroSec_CAF1 4468 119 - 1 GUGCCUGGGGGAUGGCUGCUGUUAAUUGGCUGCCACAAUUUGACUUUUUGUCGCCUAUCGGGCAAACAAAAUCAGUUCCAGGGGCAUGUCCGUGAGCGACAUCAUCAUUUCC-CCAAGAC ((.(((((((..((((.((((.....)))).))))....((((..((((((.(((.....)))..))))))))))))))))).))((((((....).)))))..........-....... ( -38.50) >DroSim_CAF1 5014 119 - 1 GUGCCUGGGGGAUGGCUGCUGUUAAUUGGCUGCCACAAUUUGACUUUUUGUCGCCUAUCGGGCAAACAAAAUCAGUUCCAGGGGCAUGUCCGUGGGCGACAUCAUCAUUUCC-CCAAGAC .....(((((((((((.((((.....)))).)))).....(((.....(((((((((.((((((..(....((.......)).)..)))))))))))))))...))).))))-))).... ( -43.60) >DroEre_CAF1 4574 119 - 1 GUGCCUGGGGAAUGGCUGCUGUUAAUUGGCUGCCACAAUUUGACUUUUUGUCGCCUAUCGGGCAAACAGAAUCAGUUCCAGGGGCAUGUCCGUGGGCGACAUCAUCAUUUCCC-CAAGAC .....(((((((((((.((((.....)))).)))).....(((.....(((((((((.((((((..(.(((....)))....)...)))))))))))))))...))).)))))-)).... ( -44.90) >DroYak_CAF1 5028 120 - 1 GUGCCCGGGGGAUGGCUGCUGUUAAUUGGCUGCCACAAUUUGACUUUUUGUCGCCUAUCGGGCAAACAGAAUCAGUUCCAGGGGCAUGUCCGUGGGCGACAUCAUCAUUUCCCCCAAGAU ......((((((((((.((((.....)))).)))).....(((.....(((((((((.((((((..(.(((....)))....)...)))))))))))))))...)))..))))))..... ( -46.80) >consensus GUGCCUGGGGGAUGGCUGCUGUUAAUUGGCUGCCACAAUUUGACUUUUUGUCGCCUAUCGGGCAAACAAAAUCAGUUCCAGGGGCAUGUCCGUGGGCGACAUCAUCAUUUCCCCCAAGAC ......((((((((((.((((.....)))).)))).....(((.....(((((((((.((((((..(....((.......)).)..)))))))))))))))...)))..))))))..... (-36.84 = -38.04 + 1.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:04:46 2006