
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 21,032,274 – 21,032,392 |
| Length | 118 |
| Max. P | 0.992079 |

| Location | 21,032,274 – 21,032,392 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.14 |
| Mean single sequence MFE | -36.32 |
| Consensus MFE | -29.78 |
| Energy contribution | -30.34 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.82 |
| SVM decision value | 2.31 |
| SVM RNA-class probability | 0.992079 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
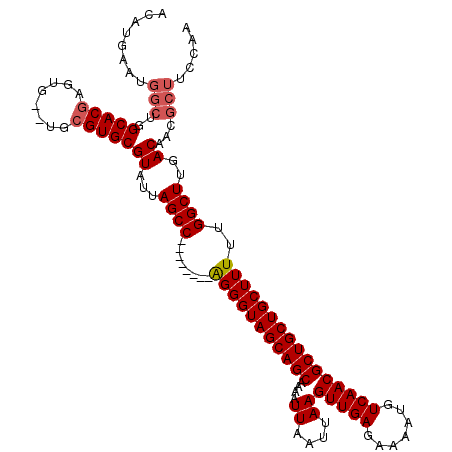
>X_DroMel_CAF1 21032274 118 + 22224390 AUAUGAAUGGCUGGCACAAGUG--UGCGUGCGUAUUAGCCGAGGUAGCAGGGUAGCAGCAAAAUUAAUUAAGUUGAGAAAAUGUCAACGCUGCUGCUUUUUGGCUUGACAACGCUUCUCA ...(((..(((..((((....)--)))((..((...((((((((((((((.((.(((......(((((...))))).....)))..)).))))))))..))))))..)).)))))..))) ( -37.60) >DroSec_CAF1 3041 110 + 1 ACAUGAAUGGCUGGCACGAGUG--UGCGUGCGUAUUAGCC--------AGGGUAGCAGCAAAAUUAAUUAAGUUGAGAAAAUGUCAACGCUGCUGCUUUUUGGCUUGACAACGCUUCCAA ...((...(((..(((((....--..)))))((...((((--------(((((((((((....((....))(((((.......))))))))))))))..))))))..))...)))..)). ( -36.80) >DroSim_CAF1 3571 110 + 1 ACAUGAAUGGCUGGCACGAGUG--UGCGUGCGUAUUAGCC--------AGGGUAGCAGCAAAAUUAAUUAAGUUGAGAAAAUGUCAACGCUGCUGCUUUUUGGCUUGACAACGCUUCCAA ...((...(((..(((((....--..)))))((...((((--------(((((((((((....((....))(((((.......))))))))))))))..))))))..))...)))..)). ( -36.80) >DroEre_CAF1 3107 109 + 1 ACAUG-AUGGCUGGCACGAGUG--UGCGUGCGUAUUAGCC--------GGGGUAGCAGCAAAAUUAAUUAAGUUGAGAAAAUGUCAACGCUGCUGCUUUUUGGCUUGACAACGCUGCUCA ...((-(((((..(((((....--..)))))((...((((--------(((((((((((....((....))(((((.......))))))))))))))..))))))..))...)))).))) ( -37.20) >DroYak_CAF1 3297 104 + 1 ACAUGAAUGGCUGGCACGAGUGUGUGCGUGCGUAUUAGCC--------GGGGUAGCAGCAAAAUUAAUUAAGUUGAGAAAAUGUCAACGCUGCUGCUUUUUGGCUUGACAAC-------- .........((..(((((....)))))..))((...((((--------(((((((((((....((....))(((((.......))))))))))))))..))))))..))...-------- ( -33.20) >consensus ACAUGAAUGGCUGGCACGAGUG__UGCGUGCGUAUUAGCC________AGGGUAGCAGCAAAAUUAAUUAAGUUGAGAAAAUGUCAACGCUGCUGCUUUUUGGCUUGACAACGCUUCCAA ........(((..(((((........)))))((...((((........(((((((((((....((....))(((((.......))))))))))))))))..))))..))...)))..... (-29.78 = -30.34 + 0.56)

| Location | 21,032,274 – 21,032,392 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.14 |
| Mean single sequence MFE | -29.75 |
| Consensus MFE | -23.05 |
| Energy contribution | -22.89 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.52 |
| SVM RNA-class probability | 0.961005 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21032274 118 - 22224390 UGAGAAGCGUUGUCAAGCCAAAAAGCAGCAGCGUUGACAUUUUCUCAACUUAAUUAAUUUUGCUGCUACCCUGCUACCUCGGCUAAUACGCACGCA--CACUUGUGCCAGCCAUUCAUAU ...(((((((.....((((....((((((((((((((.......)))))..((......))))))).....)))).....))))...))))..(((--(....))))......))).... ( -28.90) >DroSec_CAF1 3041 110 - 1 UUGGAAGCGUUGUCAAGCCAAAAAGCAGCAGCGUUGACAUUUUCUCAACUUAAUUAAUUUUGCUGCUACCCU--------GGCUAAUACGCACGCA--CACUCGUGCCAGCCAUUCAUGU .(((..((((.....(((((...((((((((.(((((................))))).))))))))....)--------))))...))))..(((--(....))))...)))....... ( -31.59) >DroSim_CAF1 3571 110 - 1 UUGGAAGCGUUGUCAAGCCAAAAAGCAGCAGCGUUGACAUUUUCUCAACUUAAUUAAUUUUGCUGCUACCCU--------GGCUAAUACGCACGCA--CACUCGUGCCAGCCAUUCAUGU .(((..((((.....(((((...((((((((.(((((................))))).))))))))....)--------))))...))))..(((--(....))))...)))....... ( -31.59) >DroEre_CAF1 3107 109 - 1 UGAGCAGCGUUGUCAAGCCAAAAAGCAGCAGCGUUGACAUUUUCUCAACUUAAUUAAUUUUGCUGCUACCCC--------GGCUAAUACGCACGCA--CACUCGUGCCAGCCAU-CAUGU ..(((((((((((...((......)).)))))(((((.......)))))............)))))).....--------((((.....(((((..--....))))).))))..-..... ( -30.80) >DroYak_CAF1 3297 104 - 1 --------GUUGUCAAGCCAAAAAGCAGCAGCGUUGACAUUUUCUCAACUUAAUUAAUUUUGCUGCUACCCC--------GGCUAAUACGCACGCACACACUCGUGCCAGCCAUUCAUGU --------((((...((((....((((((((.(((((................))))).)))))))).....--------)))).....(((((........)))))))))......... ( -25.89) >consensus UGAGAAGCGUUGUCAAGCCAAAAAGCAGCAGCGUUGACAUUUUCUCAACUUAAUUAAUUUUGCUGCUACCCU________GGCUAAUACGCACGCA__CACUCGUGCCAGCCAUUCAUGU .......................((((((((.(((((................))))).)))))))).............((((.....(((((........))))).))))........ (-23.05 = -22.89 + -0.16)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:04:42 2006