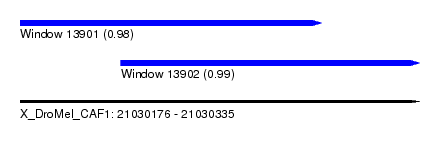
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 21,030,176 – 21,030,335 |
| Length | 159 |
| Max. P | 0.989784 |

| Location | 21,030,176 – 21,030,296 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.33 |
| Mean single sequence MFE | -38.06 |
| Consensus MFE | -34.84 |
| Energy contribution | -35.00 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.96 |
| SVM RNA-class probability | 0.983821 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21030176 120 + 22224390 GCUGGGGCGGAAAUGAAUGUUAUUUAUUUAUGUCGCUGCAACGUGGUUGUUGGCCCUGUUAUUUGUUGUACUUAUUAGCAGUACAUAAGGAUAACUAUCAGCCGGCGGCGGGUGCACUUU ((((.(((((..(((((((.....))))))).))))).))((.(.(((((((((...(((((((..((((((.......))))))...))))))).....))))))))).)))))..... ( -37.70) >DroSec_CAF1 1016 120 + 1 GCUGGGGCGGAAAUGAAUGUUAUUUAUUUAUGUCGCUGAAACGUGGUUGUUGGCCCUGUUAUUUGUUGUACUUAUUAGCAGUACAUAAGGAUAACUAUCAGCCGGCGGCGGGUGCACUUU ((...(((((..(((((((.....))))))).)))))...((.(.(((((((((...(((((((..((((((.......))))))...))))))).....))))))))).)))))..... ( -34.80) >DroSim_CAF1 1522 120 + 1 GCUGGGGCGGAAAUGAAUGUUAUUUAUUUAUGUCGCUGCAACGUGGUUGUUGGCCCUGUUAUUUGUUGUACUUAUUAGCAGUACAUAAGGAUAACUAUCAGCCGGCGGCGGGUGCACUUU ((((.(((((..(((((((.....))))))).))))).))((.(.(((((((((...(((((((..((((((.......))))))...))))))).....))))))))).)))))..... ( -37.70) >DroEre_CAF1 1133 119 + 1 GCUG-GGCGGAAAUGAAUGUUAUUUAUUUAUGUCGCUGCGGCGUGGUUGUUGGCCCCGUUAUUUGUUGUACUUAUUAGCAGUACAUAAGGAUAACUAUCAGCCGCCGGCGGUUGCACUUU ((((-(((((..(((((((.....))))))).)))))((((((.(((.....))).)(((((((..((((((.......))))))...))))))).....)))))))))........... ( -38.20) >DroYak_CAF1 1222 120 + 1 GCUGGGGCGGAAAUGAAUGUUAUUUAUUUAUGUCGCUGCGGCGUGGUUGUUGGCCCUGUUAUUUGUUGUACUUAUUAGCAGUACAUAAGGAUAACUAUCAGCCGGCGGCGGGUGCACUUU ((((.(((((..(((((((.....))))))).))))).))))((((((((((((...(((((((..((((((.......))))))...))))))).....))))))))).....)))... ( -41.90) >consensus GCUGGGGCGGAAAUGAAUGUUAUUUAUUUAUGUCGCUGCAACGUGGUUGUUGGCCCUGUUAUUUGUUGUACUUAUUAGCAGUACAUAAGGAUAACUAUCAGCCGGCGGCGGGUGCACUUU ((((.(((((..(((((((.....))))))).))))).)).....(((((((((...(((((((..((((((.......))))))...))))))).....)))))))))....))..... (-34.84 = -35.00 + 0.16)

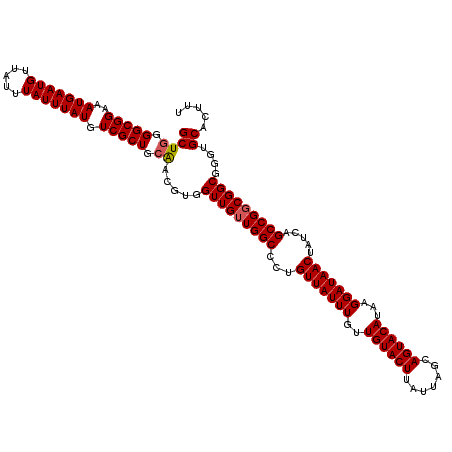
| Location | 21,030,216 – 21,030,335 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.98 |
| Mean single sequence MFE | -37.44 |
| Consensus MFE | -36.24 |
| Energy contribution | -36.00 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.18 |
| SVM RNA-class probability | 0.989784 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21030216 119 + 22224390 ACGUGGUUGUUGGCCCUGUUAUUUGUUGUACUUAUUAGCAGUACAUAAGGAUAACUAUCAGCCGGCGGCGGGUGCACUUUUAUCCGGCAAAGUUUC-CCUGGUAAUAAAGUUUACCCCAG .....(((((((((...(((((((..((((((.......))))))...))))))).....)))))))))(((((.(((((...((((.........-.))))....))))).)))))... ( -38.40) >DroSec_CAF1 1056 119 + 1 ACGUGGUUGUUGGCCCUGUUAUUUGUUGUACUUAUUAGCAGUACAUAAGGAUAACUAUCAGCCGGCGGCGGGUGCACUUUUACCCGGCAAAGUUUC-CCUGGUAAUAAAGUUUGCCCCAG .....(((((((((...(((((((..((((((.......))))))...))))))).....)))))))))(((((.(((((((((.((.........-)).))))..))))).)))))... ( -38.60) >DroSim_CAF1 1562 119 + 1 ACGUGGUUGUUGGCCCUGUUAUUUGUUGUACUUAUUAGCAGUACAUAAGGAUAACUAUCAGCCGGCGGCGGGUGCACUUUUACCCGGCAAAGUUUC-CCUGGUAAUAAAGUUUGCCCCAG .....(((((((((...(((((((..((((((.......))))))...))))))).....)))))))))(((((.(((((((((.((.........-)).))))..))))).)))))... ( -38.60) >DroEre_CAF1 1172 119 + 1 GCGUGGUUGUUGGCCCCGUUAUUUGUUGUACUUAUUAGCAGUACAUAAGGAUAACUAUCAGCCGCCGGCGGUUGCACUUUUACCCGGCAAAGUUUU-CCUGGUAAUAAAGUUUGCUCCAG ((.((((.(((((....(((((((..((((((.......))))))...)))))))..))))).))))))((..(((((((((((.((.((....))-)).))))..)))))..)).)).. ( -32.40) >DroYak_CAF1 1262 120 + 1 GCGUGGUUGUUGGCCCUGUUAUUUGUUGUACUUAUUAGCAGUACAUAAGGAUAACUAUCAGCCGGCGGCGGGUGCACUUUUGCCCGGCAAAGUUUUACCCGGUAAUAAAGUUUGCCCCAG .....(((((((((...(((((((..((((((.......))))))...))))))).....)))))))))(((((.(.((((((...)))))).).)))))(((((......))))).... ( -39.20) >consensus ACGUGGUUGUUGGCCCUGUUAUUUGUUGUACUUAUUAGCAGUACAUAAGGAUAACUAUCAGCCGGCGGCGGGUGCACUUUUACCCGGCAAAGUUUC_CCUGGUAAUAAAGUUUGCCCCAG .....(((((((((...(((((((..((((((.......))))))...))))))).....)))))))))(((((.(((((((((.((..........)).))))..))))).)))))... (-36.24 = -36.00 + -0.24)

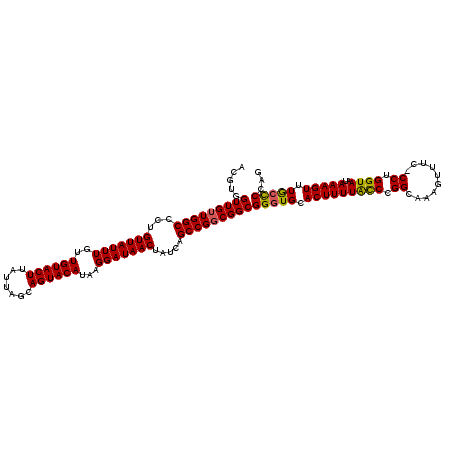
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:04:37 2006