
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 21,021,432 – 21,021,605 |
| Length | 173 |
| Max. P | 0.978727 |

| Location | 21,021,432 – 21,021,545 |
|---|---|
| Length | 113 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 76.42 |
| Mean single sequence MFE | -20.47 |
| Consensus MFE | -13.15 |
| Energy contribution | -13.27 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.694130 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21021432 113 + 22224390 ACAAUAGGUCAGUCAAUAGCGUUAUAUUAUU-UGUAUAUUUCAGAAAAUUAUUAUUUCUUGAAUUGUAUUCAAUGCAACGUGUUAUUUCUCGUCUCUGAGAGGCUAUUGUUAUC (((((((...........(((((.((((...-.(((((.((((..((((....))))..)))).)))))..)))).)))))....(((((((....)))))))))))))).... ( -23.20) >DroSec_CAF1 8182 91 + 1 GCAAUAGGCCAGU----AACGACAUAUAGUUUUUUAUAUUUCAGAAAA-UAUUAUUUCUUGAAUUGCU------------------UUCUCGUCUCUGCGAGGCUAUUGUAAUC (((((((.((.((----(..(((((((((....))))))((((((((.-.....)))).)))).....------------------.....)))..)))..))))))))).... ( -19.60) >DroSim_CAF1 8143 90 + 1 GCAAUAGGCCAGU----AACGCCACAUUAUU-UGUAUAUUUCAGAAAA-UAUUAUUUCUUGAAUUGCA------------------UUCUCGUCUCUGCGAGGCUAUUGUAAUC (((((((......----..............-((((...((((((((.-.....)))).)))).))))------------------.((((((....))))))))))))).... ( -18.60) >consensus GCAAUAGGCCAGU____AACGACAUAUUAUU_UGUAUAUUUCAGAAAA_UAUUAUUUCUUGAAUUGCA__________________UUCUCGUCUCUGCGAGGCUAUUGUAAUC (((((((................................((((((((.......)))).))))........................(((((......)))))))))))).... (-13.15 = -13.27 + 0.11)

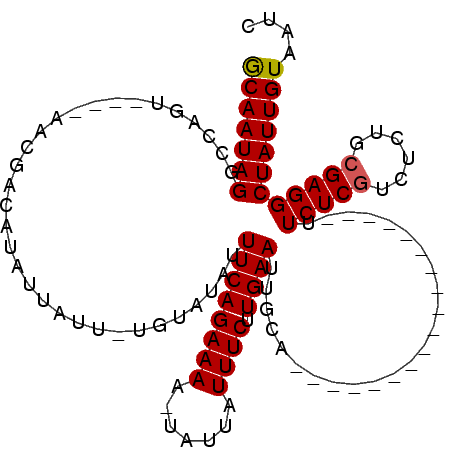
| Location | 21,021,467 – 21,021,582 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 84.05 |
| Mean single sequence MFE | -26.20 |
| Consensus MFE | -24.55 |
| Energy contribution | -24.33 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.82 |
| SVM RNA-class probability | 0.978727 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21021467 115 - 22224390 AUUUCUGGGGGCGCAAAAGCUGACUUCGGGCGCCGAAGAUAACAAUAGCCUCUCAGAGACGAGAAAUAACACGUUGCAUUGAAUACAAUUCAAGAAAUAAUAAUUUUCUGAAAUA .(((((((((((((....))...((((((...)))))).........))).))))))))..(((((......(((...((((((...))))))..)))......)))))...... ( -29.20) >DroSec_CAF1 8214 96 - 1 AUUUCUGGGGGCACAAGAGCUGACUUCGGGCGCCGAAGAUUACAAUAGCCUCGCAGAGACGAGAA------------------AGCAAUUCAAGAAAUAAUA-UUUUCUGAAAUA .(((((((((((.((.....)).((((((...)))))).........))))).))))))......------------------.....((((.((((.....-.))))))))... ( -25.70) >DroSim_CAF1 8174 96 - 1 AUUUCUGGGGGCACAAGAGCUGACUUCGGGCGUCGAAGAUUACAAUAGCCUCGCAGAGACGAGAA------------------UGCAAUUCAAGAAAUAAUA-UUUUCUGAAAUA .(((((((((((.((.....)).(((((.....))))).........))))).))))))......------------------.....((((.((((.....-.))))))))... ( -23.70) >consensus AUUUCUGGGGGCACAAGAGCUGACUUCGGGCGCCGAAGAUUACAAUAGCCUCGCAGAGACGAGAA__________________UGCAAUUCAAGAAAUAAUA_UUUUCUGAAAUA .(((((((((((.((.....)).((((((...)))))).........))))).)))))).............................((((.((((.......))))))))... (-24.55 = -24.33 + -0.22)



| Location | 21,021,505 – 21,021,605 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.07 |
| Mean single sequence MFE | -30.00 |
| Consensus MFE | -21.02 |
| Energy contribution | -21.34 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.70 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.528680 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21021505 100 - 22224390 UUAUUUCGGGUUAAAUCCAACGA--------------------AUUUCUGGGGGCGCAAAAGCUGACUUCGGGCGCCGAAGAUAACAAUAGCCUCUCAGAGACGAGAAAUAACACGUUGC (((((((((((...))))..((.--------------------.(((((((((((((....))...((((((...)))))).........))).)))))))))).)))))))........ ( -30.90) >DroSec_CAF1 8245 88 - 1 UUAUUUCGGGUUGAUUCCAACAA--------------------AUUUCUGGGGGCACAAGAGCUGACUUCGGGCGCCGAAGAUUACAAUAGCCUCGCAGAGACGAGAA------------ ...(((((.((((....))))..--------------------.(((((((((((.((.....)).((((((...)))))).........))))).))))))))))).------------ ( -28.80) >DroSim_CAF1 8205 88 - 1 UUAUUUCGGGUUGAUUCCAACAA--------------------AUUUCUGGGGGCACAAGAGCUGACUUCGGGCGUCGAAGAUUACAAUAGCCUCGCAGAGACGAGAA------------ ...(((((.((((....))))..--------------------.(((((((((((.((.....)).(((((.....))))).........))))).))))))))))).------------ ( -26.80) >DroEre_CAF1 5041 110 - 1 UUUUUGCGGGUUAAUUGCAACGAUUUGUGGCCAGAAGACGGGCUUUUCUGG-GGCACAAGAACUGGCUUCGGGCGCCGAAGAUUACAAUAGCCUCGCGGAGUCGGGAAGGA--------- .((((((((((((.(((......((((((.((((((((.....)))))).)-).))))))......((((((...))))))....)))))))).)))))))..........--------- ( -34.50) >DroYak_CAF1 6274 110 - 1 UUAUUGCGGGCUAAUUCCAACGAUUUGUGACCA-AAGACGGGCAUUUCUGGGGGCACACGACCUGGCUUCGGACGCCGAAGAUUACAAUAGCCUUGCAGAGUCGAGAAAGG--------- ...((((((((((...(((......((((.((.-.(((........)))..)).)))).....)))((((((...)))))).......))))).)))))............--------- ( -29.00) >consensus UUAUUUCGGGUUAAUUCCAACGA____________________AUUUCUGGGGGCACAAGAGCUGACUUCGGGCGCCGAAGAUUACAAUAGCCUCGCAGAGACGAGAA____________ ...(((((((......))...........................((((((((((.((.....)).((((((...)))))).........))))).))))).)))))............. (-21.02 = -21.34 + 0.32)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:04:32 2006