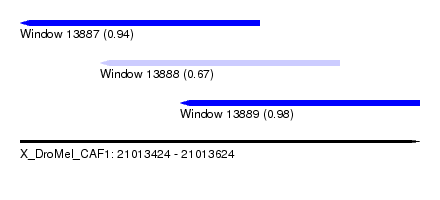
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 21,013,424 – 21,013,624 |
| Length | 200 |
| Max. P | 0.981217 |

| Location | 21,013,424 – 21,013,544 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.39 |
| Mean single sequence MFE | -49.90 |
| Consensus MFE | -48.32 |
| Energy contribution | -48.10 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.944763 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21013424 120 - 22224390 CGCCAUCAUUGGCGGAGUUCUGCUCUCAAUGAUCGAGGGCGUGGGCAUUCUGUUCACCCGCAUCUCCGCCGAGCAGUUUCGCAAUUCUGAUCCGCAGAACGAUCUAGGCAGAGCAGGCGC ((((....(((((((((....((((((.......))))))(((((...........)))))..)))))))))...(((..((..(((((.....)))))(......)))..))).)))). ( -46.90) >DroSec_CAF1 435 120 - 1 CGCCAUCAUCGGCGGAGUUCUGCUCUCAAUGAUCGAGGGCGUGGGCAUUCUGUUCACCCGCAUCUCCGCCGAGCAGUUCCGCAAUUCUGAUCCGCAGAGCGAUCUGGAAAGAGCAGGCGC ((((....(((((((((....((((((.......))))))(((((...........)))))..)))))))))((.....(((...((((.....))))))).(((....))))).)))). ( -51.80) >DroSim_CAF1 95 120 - 1 CGCCAUCAUCGGCGGAGUUCUGCUCUCAAUGAUCGAGGGCGUGGGCAUUCUGUUCACCCGCAUCUCCGCCGAGCAGUUCCGCAAUUCUGAUCCGCAGAUCGAUCUGGAAAGAGCAGGCGC ((((....(((((((((....((((((.......))))))(((((...........)))))..)))))))))((..(((((....((.((((....))))))..)))))...)).)))). ( -51.00) >consensus CGCCAUCAUCGGCGGAGUUCUGCUCUCAAUGAUCGAGGGCGUGGGCAUUCUGUUCACCCGCAUCUCCGCCGAGCAGUUCCGCAAUUCUGAUCCGCAGAACGAUCUGGAAAGAGCAGGCGC ((((....(((((((((....((((((.......))))))(((((...........)))))..)))))))))((.((((.((...........)).))))..(((....))))).)))). (-48.32 = -48.10 + -0.22)

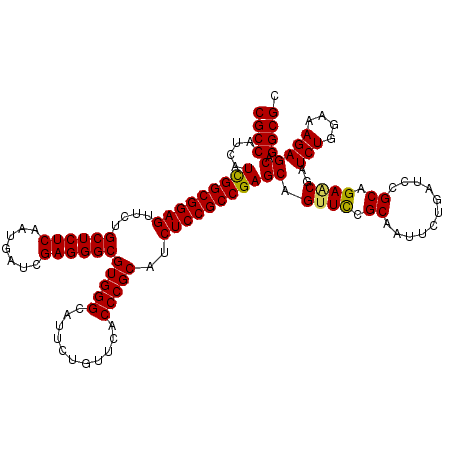

| Location | 21,013,464 – 21,013,584 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -46.60 |
| Consensus MFE | -47.26 |
| Energy contribution | -46.60 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.46 |
| Structure conservation index | 1.01 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.665592 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21013464 120 - 22224390 UUCUGUCCUCCCGAAACGGAGUAGCCGCCAUGUUCGGAAGCGCCAUCAUUGGCGGAGUUCUGCUCUCAAUGAUCGAGGGCGUGGGCAUUCUGUUCACCCGCAUCUCCGCCGAGCAGUUUC ............(((((((.....))....(((((((....((((....))))((((....((((((.......))))))(((((...........)))))..)))).)))))))))))) ( -43.60) >DroSec_CAF1 475 120 - 1 UUCUGUCCUCCCGGAACGGAGUAGCCGCCAUGUUCGGAAGCGCCAUCAUCGGCGGAGUUCUGCUCUCAAUGAUCGAGGGCGUGGGCAUUCUGUUCACCCGCAUCUCCGCCGAGCAGUUCC ............(((((((.((..(((.......)))..)).))....(((((((((....((((((.......))))))(((((...........)))))..)))))))))...))))) ( -48.10) >DroSim_CAF1 135 120 - 1 UUCUGUCCUCCCGGAACGGAGUAGCCGCCAUGUUCGGAAGCGCCAUCAUCGGCGGAGUUCUGCUCUCAAUGAUCGAGGGCGUGGGCAUUCUGUUCACCCGCAUCUCCGCCGAGCAGUUCC ............(((((((.((..(((.......)))..)).))....(((((((((....((((((.......))))))(((((...........)))))..)))))))))...))))) ( -48.10) >consensus UUCUGUCCUCCCGGAACGGAGUAGCCGCCAUGUUCGGAAGCGCCAUCAUCGGCGGAGUUCUGCUCUCAAUGAUCGAGGGCGUGGGCAUUCUGUUCACCCGCAUCUCCGCCGAGCAGUUCC ............(((((((.((..(((.......)))..)).))....(((((((((....((((((.......))))))(((((...........)))))..)))))))))...))))) (-47.26 = -46.60 + -0.66)

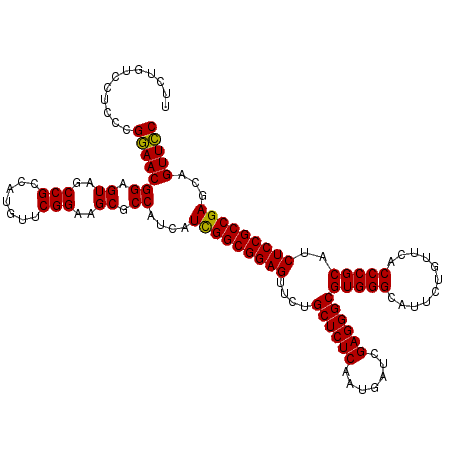
| Location | 21,013,504 – 21,013,624 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.78 |
| Mean single sequence MFE | -57.00 |
| Consensus MFE | -56.51 |
| Energy contribution | -56.40 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.99 |
| SVM decision value | 1.88 |
| SVM RNA-class probability | 0.981217 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21013504 120 - 22224390 CCGUGGAACUCGAUCAUGAGCGGAGCGAUCGCCGGUGGGAUUCUGUCCUCCCGAAACGGAGUAGCCGCCAUGUUCGGAAGCGCCAUCAUUGGCGGAGUUCUGCUCUCAAUGAUCGAGGGC .....(..((((((((((((((((((..((((((((((.(((((((.........)))))))..)))))..((.(....).)).......))))).)))))))))...)))))))))..) ( -53.60) >DroSec_CAF1 515 120 - 1 CCGUGGAACUCGAUCAUGAGCGGGGCGAUCGCCGGCGGGAUUCUGUCCUCCCGGAACGGAGUAGCCGCCAUGUUCGGAAGCGCCAUCAUCGGCGGAGUUCUGCUCUCAAUGAUCGAGGGC .....(..((((((((((((((((((..((((((((((.(((((((((....)).)))))))..)))))..((.(....).)).......))))).)))))))))...)))))))))..) ( -58.70) >DroSim_CAF1 175 120 - 1 CCGUGGAACUCGAUCAUGAGCGGGGCGAUCGCCGGCGGGAUUCUGUCCUCCCGGAACGGAGUAGCCGCCAUGUUCGGAAGCGCCAUCAUCGGCGGAGUUCUGCUCUCAAUGAUCGAGGGC .....(..((((((((((((((((((..((((((((((.(((((((((....)).)))))))..)))))..((.(....).)).......))))).)))))))))...)))))))))..) ( -58.70) >consensus CCGUGGAACUCGAUCAUGAGCGGGGCGAUCGCCGGCGGGAUUCUGUCCUCCCGGAACGGAGUAGCCGCCAUGUUCGGAAGCGCCAUCAUCGGCGGAGUUCUGCUCUCAAUGAUCGAGGGC .....(..((((((((((((((((((..((((((((((.(((((((((....)).)))))))..)))))..((.(....).)).......))))).)))))))))...)))))))))..) (-56.51 = -56.40 + -0.11)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:04:25 2006