
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 21,011,130 – 21,011,283 |
| Length | 153 |
| Max. P | 0.985898 |

| Location | 21,011,130 – 21,011,248 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 94.49 |
| Mean single sequence MFE | -42.80 |
| Consensus MFE | -37.03 |
| Energy contribution | -37.65 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.87 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.531337 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
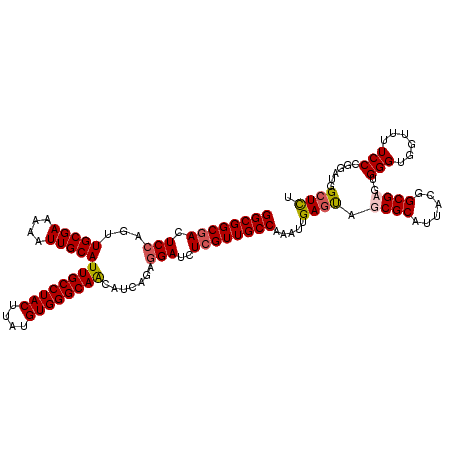
>X_DroMel_CAF1 21011130 118 - 22224390 GGCGGCGACUCCAGUUGCGAAAUAUUGCAUUGCCUACUUAUGUGGGCAACAUCAGAGGAUCUCGUUGCCAAAUUGAGUAGCGCAUUACGGCGAGUGGGUGGUUUUCCGGAAUGGCUCU ((((((((.(((...(((((....)))))((((((((....)))))))).......)))..)))))))).....((((..(((......)))..((((......)))).....)))). ( -41.80) >DroSec_CAF1 92918 118 - 1 GGCGGCGACUCCAGUUGCGAAAAAUUGCAUUGCCUACUUAUGUGGGCAACAUCAGAGGAUCUCGUUGCCAAAUUGAGUAGCGCAUUACGGCGAGUGGGUGGUUUUCCGGGAUGGCUCU ((((((((.(((...(((((....)))))((((((((....)))))))).......)))..))))))))......((.(((.(((..(((.(((.......))).)))..)))))))) ( -42.80) >DroEre_CAF1 33548 117 - 1 GGCGGCUACUCCAGUUGCGAAAAAUUGCAUUGCCUACUUAUGUGGGCAGCAGCAGA-GAUCUCGUUGCCAAAUUGAGCAGCGCAUUACGGCGCGUGGGUGGUUUUCCCGGGUGGCUUU ...(((((((((((((((((....))))..(((((((....)))))))(((((((.-...)).)))))..)))))....((((......))))..(((.......))))))))))).. ( -43.30) >DroYak_CAF1 39466 118 - 1 GGCGGCGACUCCAGUUGCGAAAAAUUGCAUUGCCUACUUAUGUGGGCAACAUCAGAGGAUCUCGUUGCCAAAUUGAGUAGCGCAUUACGGCGCGUGGGUGGUUUUCCCGGAUGGGUUU ((((((((.(((...(((((....)))))((((((((....)))))))).......)))..))))))))..........((((......)))).((((.......))))......... ( -43.30) >consensus GGCGGCGACUCCAGUUGCGAAAAAUUGCAUUGCCUACUUAUGUGGGCAACAUCAGAGGAUCUCGUUGCCAAAUUGAGUAGCGCAUUACGGCGAGUGGGUGGUUUUCCCGGAUGGCUCU ((((((((.(((...(((((....)))))((((((((....)))))))).......)))..)))))))).....((((.((((......))))..(((......)))......)))). (-37.03 = -37.65 + 0.62)

| Location | 21,011,168 – 21,011,283 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.84 |
| Mean single sequence MFE | -41.50 |
| Consensus MFE | -32.04 |
| Energy contribution | -32.35 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.58 |
| Structure conservation index | 0.77 |
| SVM decision value | 2.02 |
| SVM RNA-class probability | 0.985898 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
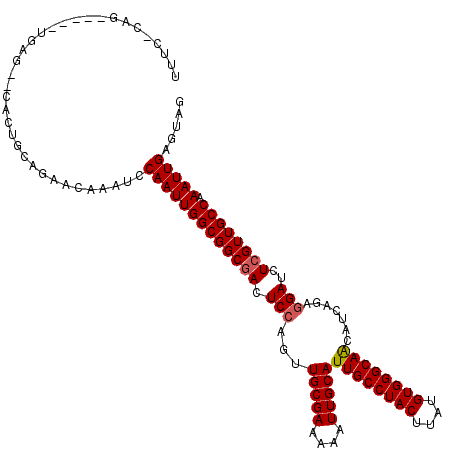
>X_DroMel_CAF1 21011168 115 - 22224390 UUUCGCAG-----UGAGAACACUGUAGAACAAAUCCAAUUGGCGGCGACUCCAGUUGCGAAAUAUUGCAUUGCCUACUUAUGUGGGCAACAUCAGAGGAUCUCGUUGCCAAAUUGAGUAG (((..(((-----((....)))))..)))......(((((((((((((.(((...(((((....)))))((((((((....)))))))).......)))..)))))))).)))))..... ( -42.80) >DroSec_CAF1 92956 113 - 1 UUUCCCAG-----UGAG--CACUGUAGAAGAAUUUCAAUUGGCGGCGACUCCAGUUGCGAAAAAUUGCAUUGCCUACUUAUGUGGGCAACAUCAGAGGAUCUCGUUGCCAAAUUGAGUAG (((..(((-----(...--.))))..)))....(((((((((((((((.(((...(((((....)))))((((((((....)))))))).......)))..)))))))).)))))))... ( -41.10) >DroEre_CAF1 33586 117 - 1 UUUUUCAGUGUGACGAG--CACUGCAGCGCAAAUCCAAUUGGCGGCUACUCCAGUUGCGAAAAAUUGCAUUGCCUACUUAUGUGGGCAGCAGCAGA-GAUCUCGUUGCCAAAUUGAGCAG ...(((((((..(((((--..((((.((((((.(((((((((.(....).))))))).))....))))..(((((((....))))))))).)))).-...)))))..)...))))))... ( -44.60) >DroYak_CAF1 39504 108 - 1 -----CAG-----UGAG--CACUGCAGAAGCAAUCCAAUUGGCGGCGACUCCAGUUGCGAAAAAUUGCAUUGCCUACUUAUGUGGGCAACAUCAGAGGAUCUCGUUGCCAAAUUGAGUAG -----(((-----(...--.))))...........(((((((((((((.(((...(((((....)))))((((((((....)))))))).......)))..)))))))).)))))..... ( -37.50) >consensus UUUC_CAG_____UGAG__CACUGCAGAACAAAUCCAAUUGGCGGCGACUCCAGUUGCGAAAAAUUGCAUUGCCUACUUAUGUGGGCAACAUCAGAGGAUCUCGUUGCCAAAUUGAGUAG ...................................(((((((((((((.(((...(((((....)))))((((((((....)))))))).......)))..)))))))).)))))..... (-32.04 = -32.35 + 0.31)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:04:20 2006