
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 21,006,339 – 21,006,487 |
| Length | 148 |
| Max. P | 0.996841 |

| Location | 21,006,339 – 21,006,433 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.09 |
| Mean single sequence MFE | -30.75 |
| Consensus MFE | -21.06 |
| Energy contribution | -20.78 |
| Covariance contribution | -0.27 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.779294 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 21006339 94 + 22224390 AUUUUGCAUAGUGCACCGUUAAAAGUAAGCCCGGGAAG----------------------CAAGGAACAG----AAUUUCCGUCAGCGGUUUAACUGCCCGGCGAUGGGCAGGCAUAAGU ..........((((((((((........((.......)----------------------)..((((...----...))))...))))))....(((((((....))))))))))).... ( -27.70) >DroSec_CAF1 88074 94 + 1 AUUUUGCAUAGUGCACCGUUAAAAAUAAGCCCGUGGAG----------------------CAAGGAACAG----AUUUUCCGUAAGCGGUUUAACUGCCCGGCGAUGGGCAGGCAUAAGU ..........((((((((((........(((....).)----------------------)..((((...----...))))...))))))....(((((((....))))))))))).... ( -29.40) >DroSim_CAF1 103576 94 + 1 AUUUUGCAUAGUGCACCGUUAAAAAUAAGCCCGGGGAG----------------------CAAGGAACAG----AAUUUCCGUAAGCGGUUUAACUGCCCGGCGAUGGGCAGGCAUAAGU ..........((((((((((........(((....).)----------------------)..((((...----...))))...))))))....(((((((....))))))))))).... ( -29.40) >DroEre_CAF1 28848 92 + 1 AUUUUGCUUAGUGCACCGUUAAA-GGAAAUCCGGGGAG----------------------CGG-GAGCAG----AAUUUCCGUAAGCGGCUUAACUGCCCGGCGAUGGGCAGGCAUAAGU .....(((((.(((.(((((...-(((((((....))(----------------------(..-..))..----..)))))...))))).....(((((((....))))))))))))))) ( -34.20) >DroYak_CAF1 34271 94 + 1 AUUUUGCAUAGUGCACCGUUAAAAGUAAAUCCGGGGAG----------------------CGGGGAACAG----AGUUUCCGUAAGCGGUUUAACUGCCCGGCGAUGGGCAGGCAUAAGU ..........((((((((((.........((....))(----------------------(((..(....----..)..)))).))))))....(((((((....))))))))))).... ( -31.40) >DroAna_CAF1 39941 120 + 1 UUUAUUCCGAGUGCACCGUUUAAAGCAAAUCUGAAAAUCUGUGAAUCUGAGAAUCUGUGGCGGGGUCCAGGACACAUUUCCGUAAGCGGUUUAACGGCCCGUCUCUGGGCUGGCAUAAGU ..........((((((((((((..........((((...((((..((((.((..(((...)))..)))))).))))))))..))))))))....(((((((....))))))))))).... ( -32.40) >consensus AUUUUGCAUAGUGCACCGUUAAAAGUAAACCCGGGGAG______________________CAAGGAACAG____AAUUUCCGUAAGCGGUUUAACUGCCCGGCGAUGGGCAGGCAUAAGU ..........((((((((((...........((((((.......................................))))))..))))))....(((((((....))))))))))).... (-21.06 = -20.78 + -0.27)


| Location | 21,006,393 – 21,006,487 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 96.09 |
| Mean single sequence MFE | -35.32 |
| Consensus MFE | -33.12 |
| Energy contribution | -33.32 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.76 |
| SVM RNA-class probability | 0.996841 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
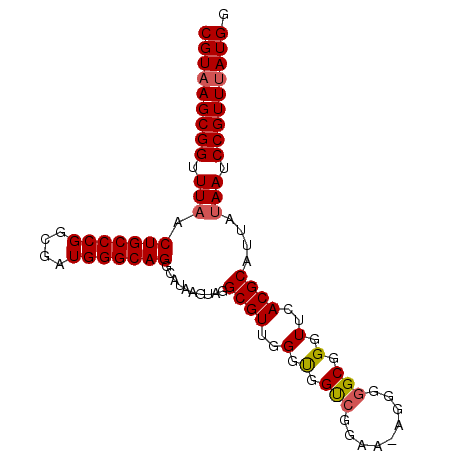
>X_DroMel_CAF1 21006393 94 + 22224390 CGUCAGCGGUUUAACUGCCCGGCGAUGGGCAGGCAUAAGUAGGCGUUGGGUGGUCGGAA-AGGGGGCGGGUUCACGCAUUAUAAUCCGUUUAUGG (((.(((((((..((..(((((((....((........))...)))))))..))..)).-.....(((......)))........))))).))). ( -31.30) >DroSec_CAF1 88128 94 + 1 CGUAAGCGGUUUAACUGCCCGGCGAUGGGCAGGCAUAAGUAGGCGUUGGGUGGUCGGAA-AGGGGGCGGGUUCACGCAUUAUAAUCCGUUUAUGG (((((((((((..((..(((((((....((........))...)))))))..))..)).-.....(((......)))........))))))))). ( -35.20) >DroSim_CAF1 103630 94 + 1 CGUAAGCGGUUUAACUGCCCGGCGAUGGGCAGGCAUAAGUAGGCGUUGGGUGGUCGGAA-AGGGGGCGGGUUCACGCAUUAUAAUCCGUUUAUGG (((((((((((..((..(((((((....((........))...)))))))..))..)).-.....(((......)))........))))))))). ( -35.20) >DroEre_CAF1 28900 95 + 1 CGUAAGCGGCUUAACUGCCCGGCGAUGGGCAGGCAUAAGUAGGCGUUGGGCGGUGGGAACAGGGGGCGGGUUCACGCAUUAAAAUCCGUUUAUGG (((((((((((((.((((((((((....((........))...))))))))))))))........(((......)))........))))))))). ( -36.30) >DroYak_CAF1 34325 95 + 1 CGUAAGCGGUUUAACUGCCCGGCGAUGGGCAGGCAUAAGUAGGCGUUGGGCGGCCGGAAAAGGGGGCAGGUUCACGCAUUAUAAUCCGUUUAUGG (((((((((.(((.(((((((....)))))))..........((((.((((.(((.........)))..))))))))....))).))))))))). ( -38.60) >consensus CGUAAGCGGUUUAACUGCCCGGCGAUGGGCAGGCAUAAGUAGGCGUUGGGUGGUCGGAA_AGGGGGCGGGUUCACGCAUUAUAAUCCGUUUAUGG (((((((((.(((.(((((((....)))))))..........((((..(.(.(((.........))).).)..))))....))).))))))))). (-33.12 = -33.32 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:04:15 2006