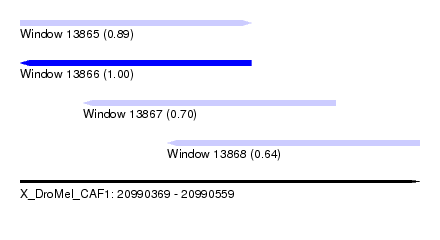
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,990,369 – 20,990,559 |
| Length | 190 |
| Max. P | 0.999725 |

| Location | 20,990,369 – 20,990,479 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.04 |
| Mean single sequence MFE | -34.08 |
| Consensus MFE | -26.52 |
| Energy contribution | -26.88 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.891519 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20990369 110 + 22224390 ----AGGAUG------AGGAUAGCGAUGAAGACAUAUGCUAAUCUUCAUCGAAGGAAAGCGCCGACAAUGGAUGGUAAUGGCGGAACCCAAAUGGGUGGAAAAUUCCUUUGUCUUUAAUG ----..((((------((((((((((((....))).)))).)))))))))(((((((..((((.((........))...))))..((((....))))......))))))).......... ( -35.70) >DroSec_CAF1 71889 114 + 1 UUCGUGGAUG------AGGAUAGCGAUGAAGACAUAUGCUAAUCUUCACCGAAGGAGAGCGCCGACAAUGGAUGGUAAUGGCGGGACCCAAAUGGGAGGAAAAUUCCUUUGUCUUUAAUG .....((.((------((((((((((((....))).)))).)))))))))(((((((..((((.((........))...))))...(((....))).......))))))).......... ( -32.50) >DroSim_CAF1 82686 114 + 1 UUCGUGGAUG------AGGAUAGCGAUGAAGACAUAUGCUAAUCUUCAUCGAAGGAGAGCGCCGACAAUGGAUGGUAAUGGCGGGACCCAAAUGGGUGGAAAAUUCCUUUGUCUUUAAUG ......((((------((((((((((((....))).)))).)))))))))(((((((..((((.((........))...))))..((((....))))......))))))).......... ( -35.50) >DroEre_CAF1 14400 114 + 1 CUCGUGGAUG------AGGAUAGCGAUGAAGACAUAUGCUAAUCUUCAUCGAAGGAAAGCGCCGACAAUGGAUGGCAAUGGCAGGACCCAAAUGGGUGGAAAAUUCCUUUGUCUUUAAUG ......((((------((((((((((((....))).)))).)))))))))(((((((...((((.(....).)))).........((((....))))......))))))).......... ( -34.30) >DroYak_CAF1 16308 120 + 1 UUCGUGGAUGAGAAUGAGGAUAGCGAUGAAGACAUAUGCUAAUCUUCAUCGAUGGAAAGCGCCGACAAUGGAUGGCAAUGGCAGGACCCAAAUGGGUGGAAAAUUCCUUUGUCUUUAAUG (((((.(((((.....(((.((((((((....))).))))).)))))))).)))))....((((.(....).))))...((((((((((....))))(((....)))))))))....... ( -32.40) >consensus UUCGUGGAUG______AGGAUAGCGAUGAAGACAUAUGCUAAUCUUCAUCGAAGGAAAGCGCCGACAAUGGAUGGUAAUGGCGGGACCCAAAUGGGUGGAAAAUUCCUUUGUCUUUAAUG ................((((((.(((((((((..........)))))))))((((((...((((.((.....))....))))...((((....))))......))))))))))))..... (-26.52 = -26.88 + 0.36)

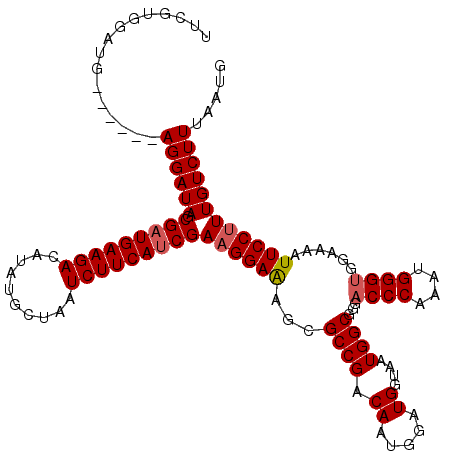
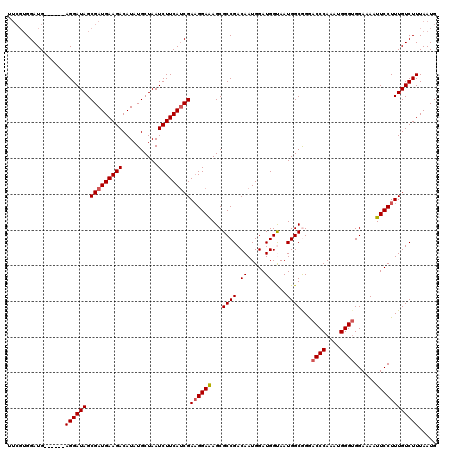
| Location | 20,990,369 – 20,990,479 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.04 |
| Mean single sequence MFE | -27.25 |
| Consensus MFE | -24.82 |
| Energy contribution | -24.86 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.91 |
| SVM decision value | 3.95 |
| SVM RNA-class probability | 0.999725 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20990369 110 - 22224390 CAUUAAAGACAAAGGAAUUUUCCACCCAUUUGGGUUCCGCCAUUACCAUCCAUUGUCGGCGCUUUCCUUCGAUGAAGAUUAGCAUAUGUCUUCAUCGCUAUCCU------CAUCCU---- ...........((((((......((((....))))..((((................))))..))))))((((((((((........)))))))))).......------......---- ( -27.29) >DroSec_CAF1 71889 114 - 1 CAUUAAAGACAAAGGAAUUUUCCUCCCAUUUGGGUCCCGCCAUUACCAUCCAUUGUCGGCGCUCUCCUUCGGUGAAGAUUAGCAUAUGUCUUCAUCGCUAUCCU------CAUCCACGAA .......(((((((((....))))(((....)))..................)))))((..........((((((((((........)))))))))).......------...))..... ( -25.25) >DroSim_CAF1 82686 114 - 1 CAUUAAAGACAAAGGAAUUUUCCACCCAUUUGGGUCCCGCCAUUACCAUCCAUUGUCGGCGCUCUCCUUCGAUGAAGAUUAGCAUAUGUCUUCAUCGCUAUCCU------CAUCCACGAA .......(((((.(((....)))((((....)))).................)))))((..........((((((((((........)))))))))).......------...))..... ( -27.15) >DroEre_CAF1 14400 114 - 1 CAUUAAAGACAAAGGAAUUUUCCACCCAUUUGGGUCCUGCCAUUGCCAUCCAUUGUCGGCGCUUUCCUUCGAUGAAGAUUAGCAUAUGUCUUCAUCGCUAUCCU------CAUCCACGAG .......(((((.(((.......((((....))))...((....))..))).)))))((..........((((((((((........)))))))))).......------...))..... ( -28.85) >DroYak_CAF1 16308 120 - 1 CAUUAAAGACAAAGGAAUUUUCCACCCAUUUGGGUCCUGCCAUUGCCAUCCAUUGUCGGCGCUUUCCAUCGAUGAAGAUUAGCAUAUGUCUUCAUCGCUAUCCUCAUUCUCAUCCACGAA .......(((((.(((.......((((....))))...((....))..))).)))))((......))..((((((((((........))))))))))....................... ( -27.70) >consensus CAUUAAAGACAAAGGAAUUUUCCACCCAUUUGGGUCCCGCCAUUACCAUCCAUUGUCGGCGCUUUCCUUCGAUGAAGAUUAGCAUAUGUCUUCAUCGCUAUCCU______CAUCCACGAA .......(((((.(((....)))((((....)))).................)))))............((((((((((........))))))))))....................... (-24.82 = -24.86 + 0.04)

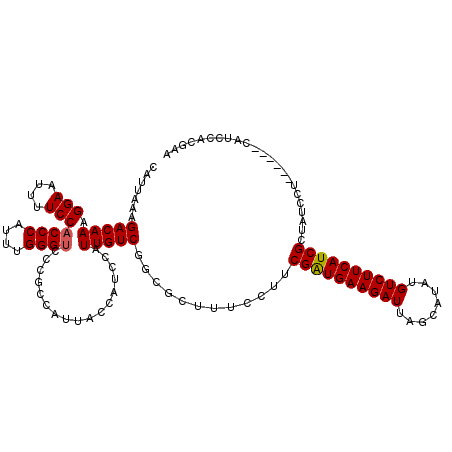
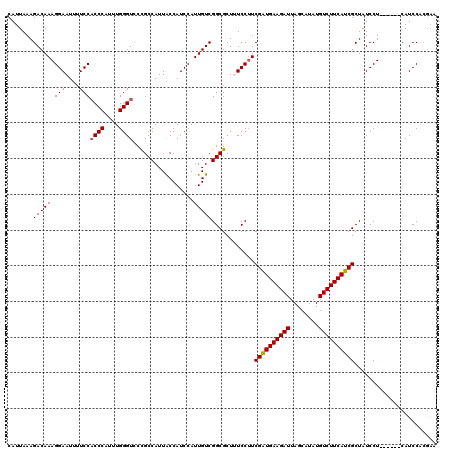
| Location | 20,990,399 – 20,990,519 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.00 |
| Mean single sequence MFE | -29.04 |
| Consensus MFE | -22.36 |
| Energy contribution | -22.60 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.702328 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20990399 120 - 22224390 UCACCCCUCAAUUCACAUUAGUUGAUGGAAAAUGCUCAUGCAUUAAAGACAAAGGAAUUUUCCACCCAUUUGGGUUCCGCCAUUACCAUCCAUUGUCGGCGCUUUCCUUCGAUGAAGAUU ....................(((((.(((((((((....)))))...(((((.(((....)))((((....)))).................)))))......)))).)))))....... ( -25.50) >DroSec_CAF1 71923 120 - 1 UCGCCCCUCCAUUCACAUUAGUUGAUGGAAAAUGCUCAUGCAUUAAAGACAAAGGAAUUUUCCUCCCAUUUGGGUCCCGCCAUUACCAUCCAUUGUCGGCGCUCUCCUUCGGUGAAGAUU .((((..((((((.((....)).)))))).(((((....)))))...(((((((((....))))(((....)))..................))))))))).(((((....).).))).. ( -30.50) >DroSim_CAF1 82720 120 - 1 UCACCCCUCCAUUCACAUUAGUUGAUGGAAAAUGCUCAUGCAUUAAAGACAAAGGAAUUUUCCACCCAUUUGGGUCCCGCCAUUACCAUCCAUUGUCGGCGCUCUCCUUCGAUGAAGAUU .....((((((((.((....)).)))))).(((((....)))))...(((((.(((....)))((((....)))).................))))))).......((((...))))... ( -25.70) >DroEre_CAF1 14434 120 - 1 UCACCCUUCCUUCCACAUUAGUUGAUGGAAAACGCUCAUGCAUUAAAGACAAAGGAAUUUUCCACCCAUUUGGGUCCUGCCAUUGCCAUCCAUUGUCGGCGCUUUCCUUCGAUGAAGAUU .........((((.......(((((.(((((.((((...........(((((.(((.......((((....))))...((....))..))).))))))))).))))).)))))))))... ( -28.41) >DroYak_CAF1 16348 120 - 1 UCAGUCUUCCUUCCACAUUAGUUGAUGGAAAAUGCUCAUGCAUUAAAGACAAAGGAAUUUUCCACCCAUUUGGGUCCUGCCAUUGCCAUCCAUUGUCGGCGCUUUCCAUCGAUGAAGAUU ..(((((((...........(((((((((((((((....)))))...(((((.(((.......((((....))))...((....))..))).)))))......))))))))))))))))) ( -35.10) >consensus UCACCCCUCCAUUCACAUUAGUUGAUGGAAAAUGCUCAUGCAUUAAAGACAAAGGAAUUUUCCACCCAUUUGGGUCCCGCCAUUACCAUCCAUUGUCGGCGCUUUCCUUCGAUGAAGAUU ....................(((((.(((.(((((....)))))...(((((.(((....)))((((....)))).................))))).......))).)))))....... (-22.36 = -22.60 + 0.24)


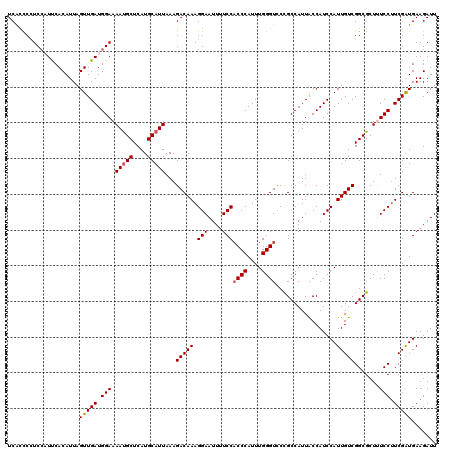
| Location | 20,990,439 – 20,990,559 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.20 |
| Mean single sequence MFE | -23.38 |
| Consensus MFE | -20.58 |
| Energy contribution | -21.34 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.641733 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20990439 120 - 22224390 GUAACAAUAUUUAAAUGACUAAGCUUUUUUUUUUUAAAUGUCACCCCUCAAUUCACAUUAGUUGAUGGAAAAUGCUCAUGCAUUAAAGACAAAGGAAUUUUCCACCCAUUUGGGUUCCGC ......................((..............((((..((.((((((......)))))).))..(((((....)))))...))))..(((.......((((....))))))))) ( -22.11) >DroSec_CAF1 71963 117 - 1 GUAGCAAUAUUUAAAUGACUAAGCUUUU---UUCAAGAUGUCGCCCCUCCAUUCACAUUAGUUGAUGGAAAAUGCUCAUGCAUUAAAGACAAAGGAAUUUUCCUCCCAUUUGGGUCCCGC ...((...(((((((((...........---.......((((.....((((((.((....)).)))))).(((((....)))))...)))).((((....))))..)))))))))...)) ( -24.50) >DroSim_CAF1 82760 117 - 1 GUAGCAAUAUUUAAAUGACUAAGCUUUU---UUCAAGAUGUCACCCCUCCAUUCACAUUAGUUGAUGGAAAAUGCUCAUGCAUUAAAGACAAAGGAAUUUUCCACCCAUUUGGGUCCCGC ...((...(((((((((...........---.......((((.....((((((.((....)).)))))).(((((....)))))...))))..(((....)))...)))))))))...)) ( -24.40) >DroEre_CAF1 14474 117 - 1 GUAGCCAUAUUUAAAUGACUAAGCAUUU---UUUAAAAUGUCACCCUUCCUUCCACAUUAGUUGAUGGAAAACGCUCAUGCAUUAAAGACAAAGGAAUUUUCCACCCAUUUGGGUCCUGC .(((.(((......))).))).(((...---.......((((....((((.((.((....)).)).))))...((....))......))))..(((....)))((((....))))..))) ( -21.60) >DroYak_CAF1 16388 117 - 1 AUAGCCAUAUUUAAAUGACUAAGCAUAU---UUUAAAAUGUCAGUCUUCCUUCCACAUUAGUUGAUGGAAAAUGCUCAUGCAUUAAAGACAAAGGAAUUUUCCACCCAUUUGGGUCCUGC .(((.(((......))).))).(((...---............(((((..(((((((.....)).)))))(((((....))))).)))))...(((....)))((((....))))..))) ( -24.30) >consensus GUAGCAAUAUUUAAAUGACUAAGCUUUU___UUUAAAAUGUCACCCCUCCAUUCACAUUAGUUGAUGGAAAAUGCUCAUGCAUUAAAGACAAAGGAAUUUUCCACCCAUUUGGGUCCCGC ...((...(((((((((.....................((((.....((((((.((....)).)))))).(((((....)))))...))))..(((....)))...)))))))))...)) (-20.58 = -21.34 + 0.76)

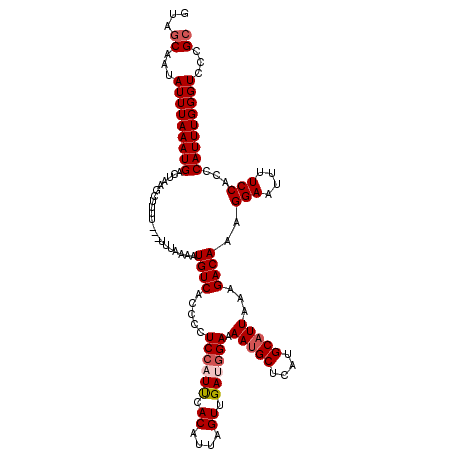
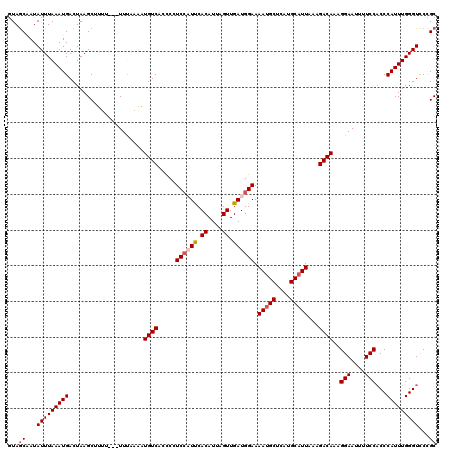
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:04:05 2006