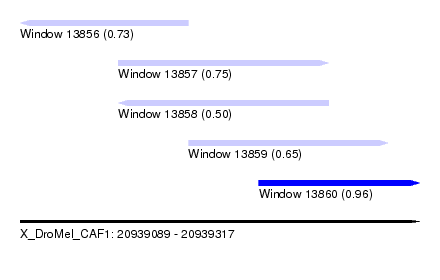
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,939,089 – 20,939,317 |
| Length | 228 |
| Max. P | 0.960087 |

| Location | 20,939,089 – 20,939,185 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 82.18 |
| Mean single sequence MFE | -31.73 |
| Consensus MFE | -19.32 |
| Energy contribution | -20.19 |
| Covariance contribution | 0.87 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.726063 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20939089 96 - 22224390 UGUAAAAUGGUUAUUUGCGAGCGCCUUAUCAGCUCCAUUCCCGC----UCGUCACGGCGUCAGGGAAUCUAAGCUGAUAAUUACAAGGUAGUCU-------GCGAAU ............(((((((..(((((((((((((..((((((..----.(((....)))...))))))...))))))))......)))).)..)-------)))))) ( -33.10) >DroSec_CAF1 11311 99 - 1 UGUAA-AUGGUUAUUUGCGAGCGGCUUAUCAGCUCCAUUCCCGCUCGCUCGUCACGGCGUCAGGGACUUUAAGCUGAUAAUUACAAGGUAGUCU-------GCGAAU .....-......(((((((..(.(((((((((((....((((...((((......))))...)))).....)))))))).......))).)..)-------)))))) ( -29.91) >DroEre_CAF1 9797 91 - 1 ----C-AUGGUUACUGGCGAGCGUCUUAUCAGCUGCAUUCCCGC----UCGCCACGGCAGUAGGGAAUUUGAGCUGAUAAUUGCAAGGUAGUCG-------CCGAAU ----.-.((((.((((.(..(((..(((((((((..((((((((----(.((....))))).))))))...))))))))).)))..).)))).)-------)))... ( -37.60) >DroYak_CAF1 10985 102 - 1 UGUAA-AUGGUUAUUUGCGAGCGGCUUAUCAGCUACAUUCCCGC----UCGUCGCGGCAGUAGGAAAUUUAAGCUGAUAAUUACAAGGUAGUCGUUAGUCUCCGAAU .((((-((....)))))).(((((((((((((((...((((..(----(.((....))))..)))).....))))))))..........)))))))........... ( -26.30) >consensus UGUAA_AUGGUUAUUUGCGAGCGGCUUAUCAGCUCCAUUCCCGC____UCGUCACGGCAGCAGGGAAUUUAAGCUGAUAAUUACAAGGUAGUCG_______CCGAAU ........(..(((((.........(((((((((..((((((.......(((....)))...))))))...))))))))).....)))))..).............. (-19.32 = -20.19 + 0.87)



| Location | 20,939,145 – 20,939,265 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.45 |
| Mean single sequence MFE | -34.20 |
| Consensus MFE | -23.66 |
| Energy contribution | -24.48 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.745932 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20939145 120 + 22224390 GAAUGGAGCUGAUAAGGCGCUCGCAAAUAACCAUUUUACACCAUGUACGACUAUAUGCAUAGUAUAUUUGGCCCAGUUUUAUGCCGGCGGGAAACUAGAGCGAUCGGUUCACGCAACAUU .....((((((((....(((((.((((((.......(((.....)))..(((((....))))).))))))(((..((.....)).)))((....)).))))))))))))).......... ( -29.30) >DroSec_CAF1 11371 119 + 1 GAAUGGAGCUGAUAAGCCGCUCGCAAAUAACCAU-UUACACCAUCUACGACUAUGUACAUAGUAUAUUUGGCCCAGUUUUCUGCCGGCGGGAAACUGGAACGAUCGGUUCACGCAACAUU .((((..(((((...((((.(((...........-.....(((......(((((....))))).....))).(((((((((((....)))))))))))..))).))))))).))..)))) ( -36.40) >DroEre_CAF1 9853 102 + 1 GAAUGCAGCUGAUAAGACGCUCGCCAGUAACCAU-G-----CG------AG------UAUAGUAUAGUUGGCCCAGUUUUCUGCCGGCGGGAAACUGGAACGAUCGGUUCACGCCACAUU ((((.((((((.((..(..(((((..........-)-----))------))------..)..))))))))..(((((((((((....)))))))))))........)))).......... ( -32.50) >DroYak_CAF1 11048 109 + 1 GAAUGUAGCUGAUAAGCCGCUCGCAAAUAACCAU-UUACACCA------AG----UAUAUAGUAUAGUUGGCCCAGUUUUCCGCCGGCGGGAAACUGGAACGAUCGGUUCACGCAACAUU .(((((.(((((...((((.(((...........-.....(((------(.----((((...)))).)))).(((((((((((....)))))))))))..))).))))))).)).))))) ( -38.60) >consensus GAAUGGAGCUGAUAAGCCGCUCGCAAAUAACCAU_UUACACCA______AC____UACAUAGUAUAGUUGGCCCAGUUUUCUGCCGGCGGGAAACUGGAACGAUCGGUUCACGCAACAUU .((((..(((((...((((.(((.......(((...................................))).(((((((((((....)))))))))))..))).))))))).))..)))) (-23.66 = -24.48 + 0.81)

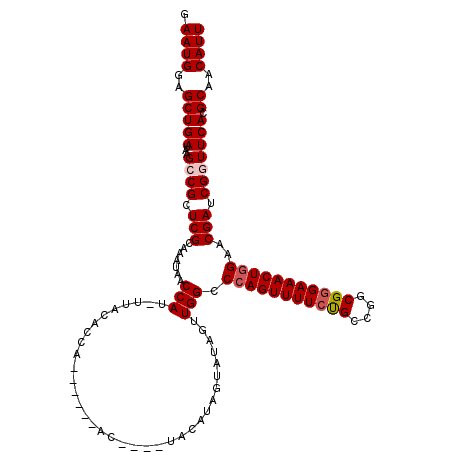

| Location | 20,939,145 – 20,939,265 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.45 |
| Mean single sequence MFE | -33.67 |
| Consensus MFE | -22.72 |
| Energy contribution | -25.09 |
| Covariance contribution | 2.37 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.67 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20939145 120 - 22224390 AAUGUUGCGUGAACCGAUCGCUCUAGUUUCCCGCCGGCAUAAAACUGGGCCAAAUAUACUAUGCAUAUAGUCGUACAUGGUGUAAAAUGGUUAUUUGCGAGCGCCUUAUCAGCUCCAUUC ...((((.((((..((.((((.(((((((((....))....)))))))((((..(((((((((.((......)).)))))))))...)))).....)))).))..))))))))....... ( -28.90) >DroSec_CAF1 11371 119 - 1 AAUGUUGCGUGAACCGAUCGUUCCAGUUUCCCGCCGGCAGAAAACUGGGCCAAAUAUACUAUGUACAUAGUCGUAGAUGGUGUAA-AUGGUUAUUUGCGAGCGGCUUAUCAGCUCCAUUC ...((((.((((.(((.((((.(((((((.(.(....).).)))))))((((..((((((((.(((......))).)))))))).-.)))).....)))).))).))))))))....... ( -37.20) >DroEre_CAF1 9853 102 - 1 AAUGUGGCGUGAACCGAUCGUUCCAGUUUCCCGCCGGCAGAAAACUGGGCCAACUAUACUAUA------CU------CG-----C-AUGGUUACUGGCGAGCGUCUUAUCAGCUGCAUUC ((((..((.(((...((.(((((((((((.(.(....).).)))))).((((.....(((((.------..------..-----.-)))))...)))))))))))...)))))..)))). ( -29.70) >DroYak_CAF1 11048 109 - 1 AAUGUUGCGUGAACCGAUCGUUCCAGUUUCCCGCCGGCGGAAAACUGGGCCAACUAUACUAUAUA----CU------UGGUGUAA-AUGGUUAUUUGCGAGCGGCUUAUCAGCUACAUUC (((((.((((((.(((.((((.(((((((.(((....))).)))))))((((..(((((((....----..------))))))).-.)))).....)))).))).))))..)).))))). ( -38.90) >consensus AAUGUUGCGUGAACCGAUCGUUCCAGUUUCCCGCCGGCAGAAAACUGGGCCAAAUAUACUAUAUA____CU______UGGUGUAA_AUGGUUAUUUGCGAGCGGCUUAUCAGCUCCAUUC ((((..((((((.(((.((((.(((((((.(.(....).).)))))))((((..((((((((..............))))))))...)))).....)))).))).))))..))..)))). (-22.72 = -25.09 + 2.37)



| Location | 20,939,185 – 20,939,299 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 87.24 |
| Mean single sequence MFE | -32.55 |
| Consensus MFE | -30.06 |
| Energy contribution | -30.00 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.654923 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20939185 114 + 22224390 CCAUGUACGACUAUAUGCAUAGUAUAUUUGGCCCAGUUUUAUGCCGGCGGGAAACUAGAGCGAUCGGUUCACGCAACAUUAGCCGGGGAUCAAUCAAGCAUUAGAUAAUCUCCG (((.(((..(((((....))))).))).)))..............(((((....)).((((.....))))...........)))((((((..(((........))).)))))). ( -25.10) >DroSec_CAF1 11410 114 + 1 CCAUCUACGACUAUGUACAUAGUAUAUUUGGCCCAGUUUUCUGCCGGCGGGAAACUGGAACGAUCGGUUCACGCAACAUUAGCCGGGGAUCAAUCAAGCAUUAGAUAAUCCCCG .........(((((....)))))......(((.((((((((((....))))))))))((((.....))))...........)))((((((..(((........))).)))))). ( -35.00) >DroEre_CAF1 9888 101 + 1 -CG------AG------UAUAGUAUAGUUGGCCCAGUUUUCUGCCGGCGGGAAACUGGAACGAUCGGUUCACGCCACAUUAGCCGGGGAUCAAUCAAGCAUUAGAUAAUCCCCG -..------..------............((((((((((((((....))))))))))).......(((....)))......)))((((((..(((........))).)))))). ( -34.30) >DroYak_CAF1 11087 104 + 1 CCA------AG----UAUAUAGUAUAGUUGGCCCAGUUUUCCGCCGGCGGGAAACUGGAACGAUCGGUUCACGCAACAUUAGCCGGGGAUCAAUCAAGCAUUAGAUAAUCCCCG (((------(.----((((...)))).)))).(((((((((((....))))))))))).......((((...........))))((((((..(((........))).)))))). ( -35.80) >consensus CCA______AC____UACAUAGUAUAGUUGGCCCAGUUUUCUGCCGGCGGGAAACUGGAACGAUCGGUUCACGCAACAUUAGCCGGGGAUCAAUCAAGCAUUAGAUAAUCCCCG .............................(((.((((((((((....))))))))))((((.....))))...........)))((((((..(((........))).)))))). (-30.06 = -30.00 + -0.06)



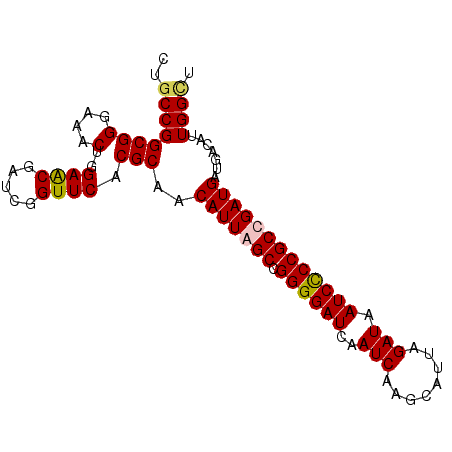
| Location | 20,939,225 – 20,939,317 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 95.11 |
| Mean single sequence MFE | -32.02 |
| Consensus MFE | -30.00 |
| Energy contribution | -30.25 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.960087 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20939225 92 + 22224390 AUGCCGGCGGGAAACUAGAGCGAUCGGUUCACGCAACAUUAGCCGGGGAUCAAUCAAGCAUUAGAUAAUCUCCGCUGAUGAUGACAUUGGUU ..((((((((....)..((((.....)))).)))..(((((((.((((((..(((........))).))))))))))))).......)))). ( -29.40) >DroSec_CAF1 11450 92 + 1 CUGCCGGCGGGAAACUGGAACGAUCGGUUCACGCAACAUUAGCCGGGGAUCAAUCAAGCAUUAGAUAAUCCCCGCUGAUGAUGACAUUGGGU ..(((.((((....)..((((.....)))).)))..(((((((.((((((..(((........))).))))))))))))).........))) ( -31.90) >DroEre_CAF1 9915 92 + 1 CUGCCGGCGGGAAACUGGAACGAUCGGUUCACGCCACAUUAGCCGGGGAUCAAUCAAGCAUUAGAUAAUCCCCGCCGAUGAUGACAUUGGCU ..((((((((....)..((((.....)))).)))).(((..((.((((((..(((........))).))))))))..)))........))). ( -34.40) >DroYak_CAF1 11117 92 + 1 CCGCCGGCGGGAAACUGGAACGAUCGGUUCACGCAACAUUAGCCGGGGAUCAAUCAAGCAUUAGAUAAUCCCCGCCGAUGAUGACAUUGGCU .....(((((....)).((((.....))))...........)))((((((..(((........))).))))))(((((((....))))))). ( -32.40) >consensus CUGCCGGCGGGAAACUGGAACGAUCGGUUCACGCAACAUUAGCCGGGGAUCAAUCAAGCAUUAGAUAAUCCCCGCCGAUGAUGACAUUGGCU ..((((((((....)..((((.....)))).)))..(((((((.((((((..(((........))).))))))))))))).......)))). (-30.00 = -30.25 + 0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:03:58 2006