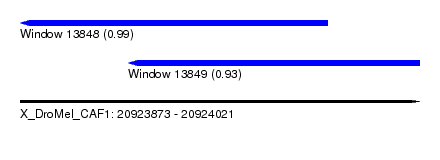
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,923,873 – 20,924,021 |
| Length | 148 |
| Max. P | 0.985473 |

| Location | 20,923,873 – 20,923,987 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 87.17 |
| Mean single sequence MFE | -43.08 |
| Consensus MFE | -34.60 |
| Energy contribution | -35.92 |
| Covariance contribution | 1.32 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.80 |
| SVM decision value | 2.01 |
| SVM RNA-class probability | 0.985473 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20923873 114 - 22224390 UUGGCAUUAGCUCCGCCAGGUGGAGUUCCAGUUGCAGGCACCUUCGUGCCAGUGACU---CACCUUCCUGCCCACUUGCUUGGGGCUUCUGAACACCCUGUUCCUCUGGUCCCAGUU ..((((..((((((((...))))))))..(((..(.(((((....))))).)..)))---........)))).......((((((((...((((.....))))....)))))))).. ( -42.00) >DroSec_CAF1 4244 114 - 1 UUGACAUUGGCUCCGCCAGGUGGAGUUCCAGUUGCAGGCACCUUUGUGCCUGUGACU---CGCCUUCCUGCCCACUUGCUUGGGGCUUCUGAACACCCUGUUCCUCUGGUCCCAGUU .......((((...))))(((((((..(.(((..(((((((....)))))))..)))---.)..)))).))).......((((((((...((((.....))))....)))))))).. ( -44.80) >DroSim_CAF1 4249 114 - 1 UUGGCAUUGGCUCCGCCAGGUGGAGUUCCAGUUGCAGGCACCUUUGUGCCUGUGACU---CGCCUUCCUGCCCACUUGCUUGGGGCUUCUGAACACCCUGUUCCUCUGGUCCCAGUU ..((((.((((...))))(((((((..(.(((..(((((((....)))))))..)))---.)..)))).)))....))))(((((((...((((.....))))....)))))))... ( -47.60) >DroEre_CAF1 4429 117 - 1 UUGGCACUGGCUCCGCCAGGUGGAGUUCCAGUUGCAGGCGCCUUCGUGCCCGUUACUCCCCGACUUUCUGCUCACCUCCUAGCAGAUGCUGAGCCCCCUGCUCCUCUGGUCCCAGUU ..((((..(((((.((..((.(((((.......((.(((((....))))).)).))))))).....((((((........)))))).)).)))))...))))...(((....))).. ( -41.20) >DroYak_CAF1 4392 117 - 1 UUGGCAUUGGCUCCGCCAGUUGGAGUUCCAGUUGCAGGCGCCUUCGUGCCUGUGCCAUCACGACUUUCUGCCCACUUCCUCGGGGAUACAGAACACCCUGUUGCUCUGGUCCCAGUG .((((((((((((((.....)))))..))))).((((((((....))))))))))))...............((((......(((((.((((.((......)).))))))))))))) ( -39.80) >consensus UUGGCAUUGGCUCCGCCAGGUGGAGUUCCAGUUGCAGGCACCUUCGUGCCUGUGACU___CGCCUUCCUGCCCACUUGCUUGGGGCUUCUGAACACCCUGUUCCUCUGGUCCCAGUU ..((((..((((((((...))))))))...(((((((((((....)))))))))))............)))).........((((((...((((.....))))....)))))).... (-34.60 = -35.92 + 1.32)


| Location | 20,923,913 – 20,924,021 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 69.64 |
| Mean single sequence MFE | -36.19 |
| Consensus MFE | -16.00 |
| Energy contribution | -17.12 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.44 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.929095 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20923913 108 - 22224390 ------CACGUAUCCUGACCUGGGAUAUCCCUAUCAAUUGUUGGCAUUAGCUCCGCCAGGUGGAGUUCCAGUUGCAGGCACCUUCGUGCCAGU---GACU---CACCUUCCUGCCCACUU ------...(((((((.....)))))))...........((.((((..((((((((...))))))))..(((..(.(((((....))))).).---.)))---........)))).)).. ( -35.50) >DroSec_CAF1 4284 108 - 1 ------CACGUAUCCUGACCUGGGAUACCCCUAUCAAUUGUUGACAUUGGCUCCGCCAGGUGGAGUUCCAGUUGCAGGCACCUUUGUGCCUGU---GACU---CGCCUUCCUGCCCACUU ------...(((((((.....)))))))...................((((...))))(((((((..(.(((..(((((((....))))))).---.)))---.)..)))).)))..... ( -38.80) >DroSim_CAF1 4289 108 - 1 ------CACGUAUCCUGACCUGGGAUACCCCUAUCAAUUGUUGGCAUUGGCUCCGCCAGGUGGAGUUCCAGUUGCAGGCACCUUUGUGCCUGU---GACU---CGCCUUCCUGCCCACUU ------...(((((((.....)))))))...........((.((((..((((((((...))))).....(((..(((((((....))))))).---.)))---.)))....)))).)).. ( -43.10) >DroEre_CAF1 4469 99 - 1 ------CACAUAUCCCGGCCUGGC------------AUUGUUGGCACUGGCUCCGCCAGGUGGAGUUCCAGUUGCAGGCGCCUUCGUGCCCGU---UACUCCCCGACUUUCUGCUCACCU ------..........((...(((------------(..(((((((((((((((((...))))))..)))).))).(((((....)))))...---.......))))....))))..)). ( -31.50) >DroYak_CAF1 4432 99 - 1 ------CACAUAUCCCGGCCUGGC------------ACUGUUGGCAUUGGCUCCGCCAGUUGGAGUUCCAGUUGCAGGCGCCUUCGUGCCUGU---GCCAUCACGACUUUCUGCCCACUU ------.........((...((((------------((....(((((.(((.((..((..(((....)))..))..)).)))...))))).))---))))...))............... ( -33.90) >DroAna_CAF1 4165 102 - 1 CGAUCCUGGAUAUCCUGCUGUGGGCUAUCCCUACCAAGUGGUGGAGGUGACUGCAUCGCGU------------CCUGGCGGCUGUAUGCCUGUAGAGUCU---CGUUAUCCUGCCAA--- .......(((((.....(((..((((((..(..(((.((((((.((....)).))))))..------------..)))..)..))).)))..))).....---...)))))......--- ( -34.32) >consensus ______CACAUAUCCUGACCUGGGAUA_CCCUAUCAAUUGUUGGCAUUGGCUCCGCCAGGUGGAGUUCCAGUUGCAGGCACCUUCGUGCCUGU___GACU___CGCCUUCCUGCCCACUU .............((......))................((.((((..((((((((...)))))))).........(((((....))))).....................)))).)).. (-16.00 = -17.12 + 1.11)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:03:48 2006