| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 2,376,163 – 2,376,264 |
| Length | 101 |
| Max. P | 0.599842 |

| Location | 2,376,163 – 2,376,264 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 67.40 |
| Mean single sequence MFE | -19.29 |
| Consensus MFE | -7.88 |
| Energy contribution | -7.60 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.41 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.599842 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
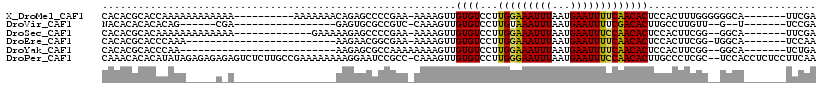
>X_DroMel_CAF1 2376163 101 - 22224390 CACACGCACCAAAAAAAAAAAA----------AAAAAAACAGAGCCCCGAA-AAAAGUUGUGUCCUUGGAAAUUUAAUGAAUUUUCAACACUCCACUUUGGGGGGCA-------UUCGA ......................----------...........(((((...-.(((((.((((...(((((((((...)))))))))))))...)))))..))))).-------..... ( -22.10) >DroVir_CAF1 38357 84 - 1 NACACACACACAG------CGA-----------------GAGUGCGCCGUC-CAAAGUUGUGUCCUUGUAAAUUUAAUGAAUUUUCGACACUUGCCUUGUU--G--U-------UCCGA .........((((------(((-----------------(.((((......-....)).(((((.....((((((...))))))..)))))..))))))))--)--)-------..... ( -13.20) >DroSec_CAF1 20183 96 - 1 CACACGCACAAAAAAAAAAAAA-------------GAAAAAGAGCCCCGAA-AAAAGUUGUGUCCUUGGAAAUUUAAUGAAUUUCCAACACUCCACUUCGG--GGCA-------UUCGA ......................-------------........((((((((-.......((((...(((((((((...))))))))))))).....)))))--))).-------..... ( -26.20) >DroEre_CAF1 21611 85 - 1 CACACGCACCCAAA-------------------------AAGAACGGCGAA-AAAAGUUGUGUCCUUGGAAAUUUAAUGAAUUUUCAACACUCCACUUCGG-UGGCA-------UCCAA ....(((.......-------------------------.......)))..-.......((((...(((((((((...))))))))))))).((((....)-)))..-------..... ( -16.54) >DroYak_CAF1 21161 84 - 1 CACACGCACCCAA--------------------------AAGAGCGCCAAAAAAAAGUUGUGUCCUUGGAAAUUUAAUGAAUUUUCAACACUCCACUUCGG--GGCA-------UCUGA .............--------------------------.(((..(((........(..((((...(((((((((...)))))))))))))..).(....)--))).-------))).. ( -16.30) >DroPer_CAF1 28167 116 - 1 CAAACACACAUAUAGAGAGAGAGUCUCUUGCCGAAAAAAAAGGAAUCCGCC-CAAAGUUGUGUCCUUGGGAAUUUAAUGAAUUUCCAACACUUGCCCUCGC--UCCACCUCUCCUUCAA ..............(((.(((((......((.((.......((.......)-)...((.((((...(((((((((...)))))))))))))..))..))))--.....))))).))).. ( -21.40) >consensus CACACGCACACAAA_____A_A_________________AAGAGCGCCGAA_AAAAGUUGUGUCCUUGGAAAUUUAAUGAAUUUUCAACACUCCACUUCGG__GGCA_______UCCGA ...........................................................((((...(((((((((...)))))))))))))............................ ( -7.88 = -7.60 + -0.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:55:08 2006