| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,920,348 – 20,920,468 |
| Length | 120 |
| Max. P | 0.859062 |

| Location | 20,920,348 – 20,920,468 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.63 |
| Mean single sequence MFE | -26.45 |
| Consensus MFE | -16.41 |
| Energy contribution | -16.58 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.859062 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
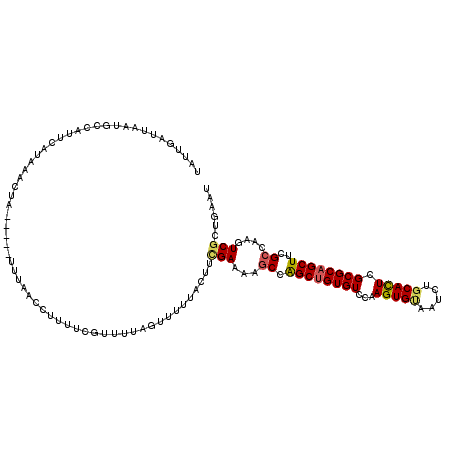
>X_DroMel_CAF1 20920348 120 + 22224390 UAUUGAUGAAUGCCAUUCAUAAGCUAUUGUAUUUAACCUUUUCGUUUUAGUUUUUACUUCGAAAAGCCAGCUGUGUCCAAGUGUAAUCUGCACUCGCGCAGCUUCGCCAAGUCGUUGAAU ..(..((((..((........................(((((((....(((....))).)))))))..((((((((...(((((.....))))).))))))))..))....))))..).. ( -28.90) >DroSec_CAF1 550 115 + 1 UAUUGAUUAAUGCUAUUCAUAAACUA-----UUUAACCUUUUCGUUUUAGUUUUUACUUCGAAAAGCCAGCUGUGUCCAAGUGUAAUCUGCACUCGCGCAGCUUCGCCAAGUCGCUGAAU ....((((...((.............-----......(((((((....(((....))).)))))))..((((((((...(((((.....))))).))))))))..))..))))....... ( -26.30) >DroSim_CAF1 558 115 + 1 UAUUAAUUAAUGCUAUUCAUAAACUA-----UUUAACCUUUUCGUUUUAGUUUUUACUUCGAAAAGCCAGCUGUGUCCAAGUGUAAUCUGCACUCGCGCAGCUUCGCCAAGUCGCUGAAU ..............(((((...(((.-----......(((((((....(((....))).)))))))..((((((((...(((((.....))))).))))))))......)))...))))) ( -25.90) >DroEre_CAF1 796 111 + 1 UUUUGUUUAAAGCUAUUAAUGAAGUA-----UUGAA----UCUUUUUUAGGUUUUACUUUGAAAAGCCAGCUGUGUCCAAGUGCAAUCUGCACUCGCGCAGCUUCGCUAAGUCGCUGAAU ..........(((.......((((((-----..(((----(((.....))))))))))))((..(((.((((((((...(((((.....))))).))))))))..)))...))))).... ( -32.00) >DroYak_CAF1 704 113 + 1 UUUUAUUUACAGCCAUUUAUAAACUA-----UGAAC--UGUUUUUUUUAGCUUUUACUUCGAAAAGCCAGCUGUGUCCAAGUGUAAUCUGCACUCGCGCAGCUUCGCUAAGUCGCUGAAU .........((((.(((((.......-----.....--...........((((((......)))))).((((((((...(((((.....))))).))))))))....))))).))))... ( -28.00) >DroPer_CAF1 421 98 + 1 UAC------GUCCCAUCC-------------UUGAUCCCUC---UGGCAGCUUUUAUUUUGAGAAACCGGCGGUGUCGAAGUGCAAUCUCCAUUCGCGCAGCUUUGCCAAGUCCCUCAAC ...------.........-------------((((..((.(---(((...(((.......)))...)))).))..)))).((((...........))))..................... ( -17.60) >consensus UAUUGAUUAAUGCCAUUCAUAAACUA_____UUUAACCUUUUCGUUUUAGUUUUUACUUCGAAAAGCCAGCUGUGUCCAAGUGUAAUCUGCACUCGCGCAGCUUCGCCAAGUCGCUGAAU ...........................................................(((...((.((((((((...(((((.....))))).))))))))..))....)))...... (-16.41 = -16.58 + 0.17)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:03:45 2006