
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,904,941 – 20,905,031 |
| Length | 90 |
| Max. P | 0.775283 |

| Location | 20,904,941 – 20,905,031 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 78.51 |
| Mean single sequence MFE | -33.35 |
| Consensus MFE | -19.44 |
| Energy contribution | -20.12 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.775283 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20904941 90 + 22224390 CGCGCC---------G---CAGCAGCAUCAGCAAGCCGGUGUAAUCAACACGAUCUCUGGCAGCAUUCCGGCCACGGGAUUUAGUUCCCUGGCCGGCAGCUU .(((((---------(---(....((....))..(((((.(..(((.....)))).))))).)).....(((((.((((......)))))))))))).)).. ( -37.10) >DroSec_CAF1 24128 90 + 1 CGCGCC---------G---CAGCAGCAUCAGCAAGCUGGCCUAAUCAGCACGAUCUCUGGCAGCAUUCCGGCCACGGGAUUUAGUUCCCUGGCCGGCAGCUU .(((((---------(---(..((((........))))(((..(((.....)))....))).)).....(((((.((((......)))))))))))).)).. ( -37.20) >DroSim_CAF1 26020 90 + 1 CGCGCC---------G---CAGCAGCAUCAGCAAGCUGGCCUAAUCAACACGAUCUCUGGCAGCAUUCCGGCCACGGGAUUUAGUUCCCUGGCCGGCAGCUU .(((((---------(---(..((((........))))(((..(((.....)))....))).)).....(((((.((((......)))))))))))).)).. ( -37.20) >DroYak_CAF1 21370 93 + 1 CGCGCC---------GCAGCAGCAGCAUCAGCAAACUGGCCUGAUCAACACGAUCUCCGGCAGCAUUCCGGCCACGGGAUUCAGUUCUCUGGCCGGCAGCUU .(((((---------((..(((..((....))...)))(((.((((.....))))...))).)).....(((((.((((......)))))))))))).)).. ( -35.30) >DroMoj_CAF1 26367 99 + 1 GAUACAGCACGAGUCG---CAAAAGCAUCUGCAUACAGGUGUCAUCAAUACGAUUUCAGGCAGCAUUCCAGCGACGGGCUUUCAGUCGCUAGCUGGCAGCUU .....(((..((((((---.....(((((((....)))))))........))))))..(.((((.....((((((.........)))))).)))).).))). ( -28.12) >DroAna_CAF1 15780 90 + 1 CGUUUC---------C---CAGCAACAGCAACAAACUGGAGUGAUCAACACCAUUUCUGGGAGUAUUCCUGCCACAGGAUUUGGUUCCAUGGCUGGCAGCUU ...(((---------(---((((....))........((((((........)))))))))))).....(((((((((((......))).))..))))))... ( -25.20) >consensus CGCGCC_________G___CAGCAGCAUCAGCAAACUGGCCUAAUCAACACGAUCUCUGGCAGCAUUCCGGCCACGGGAUUUAGUUCCCUGGCCGGCAGCUU .......................(((....((...((((....(((.....)))..))))..))...(((((((.((((......)))))))))))..))). (-19.44 = -20.12 + 0.67)



| Location | 20,904,941 – 20,905,031 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 78.51 |
| Mean single sequence MFE | -35.77 |
| Consensus MFE | -23.31 |
| Energy contribution | -23.37 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.754359 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20904941 90 - 22224390 AAGCUGCCGGCCAGGGAACUAAAUCCCGUGGCCGGAAUGCUGCCAGAGAUCGUGUUGAUUACACCGGCUUGCUGAUGCUGCUG---C---------GGCGCG .(((..(((((((((((......)))).)))))))...)))(((.......((((.....))))((((..((....)).))))---.---------)))... ( -40.30) >DroSec_CAF1 24128 90 - 1 AAGCUGCCGGCCAGGGAACUAAAUCCCGUGGCCGGAAUGCUGCCAGAGAUCGUGCUGAUUAGGCCAGCUUGCUGAUGCUGCUG---C---------GGCGCG .(((..(((((((((((......)))).)))))))...))).........(((((((....(((.(((........)))))).---)---------)))))) ( -42.70) >DroSim_CAF1 26020 90 - 1 AAGCUGCCGGCCAGGGAACUAAAUCCCGUGGCCGGAAUGCUGCCAGAGAUCGUGUUGAUUAGGCCAGCUUGCUGAUGCUGCUG---C---------GGCGCG .(((..(((((((((((......)))).)))))))...))).........(((((((....(((.(((........)))))).---)---------)))))) ( -40.20) >DroYak_CAF1 21370 93 - 1 AAGCUGCCGGCCAGAGAACUGAAUCCCGUGGCCGGAAUGCUGCCGGAGAUCGUGUUGAUCAGGCCAGUUUGCUGAUGCUGCUGCUGC---------GGCGCG .(((..((((((((.((......)).).)))))))...)))(((...((((.....)))).)))(((....)))..(((((....))---------)))... ( -36.50) >DroMoj_CAF1 26367 99 - 1 AAGCUGCCAGCUAGCGACUGAAAGCCCGUCGCUGGAAUGCUGCCUGAAAUCGUAUUGAUGACACCUGUAUGCAGAUGCUUUUG---CGACUCGUGCUGUAUC ..((.((...((((((((.(.....).))))))))...)).)).............((((.(((..((.((((((....))))---))))..)))...)))) ( -27.50) >DroAna_CAF1 15780 90 - 1 AAGCUGCCAGCCAUGGAACCAAAUCCUGUGGCAGGAAUACUCCCAGAAAUGGUGUUGAUCACUCCAGUUUGUUGCUGUUGCUG---G---------GAAACG ...((((((..((.(((......)))))))))))......((((((....((((.....)))).((((.....))))...)))---)---------)).... ( -27.40) >consensus AAGCUGCCGGCCAGGGAACUAAAUCCCGUGGCCGGAAUGCUGCCAGAGAUCGUGUUGAUUACGCCAGCUUGCUGAUGCUGCUG___C_________GGCGCG .(((..(((((((((((......)))).)))))))...)))..........(((((........((((..((....)).)))).............))))). (-23.31 = -23.37 + 0.06)

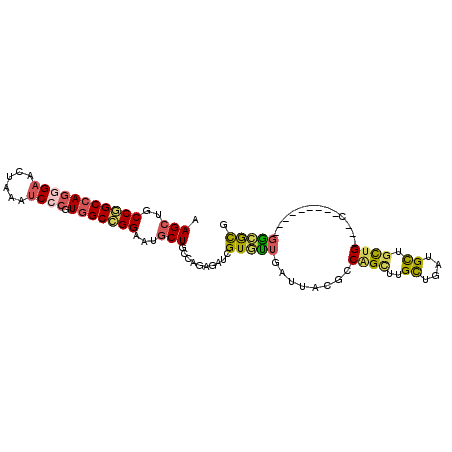
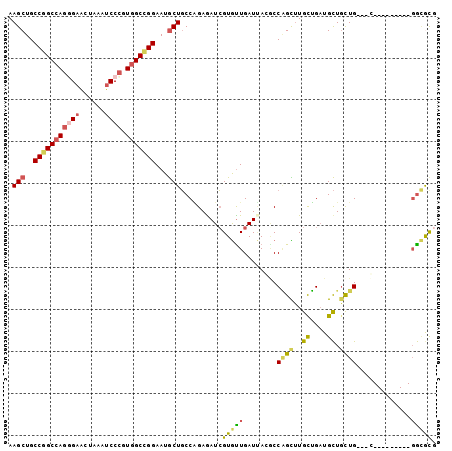
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:03:39 2006