
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,902,779 – 20,902,921 |
| Length | 142 |
| Max. P | 0.814575 |

| Location | 20,902,779 – 20,902,883 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 81.40 |
| Mean single sequence MFE | -22.07 |
| Consensus MFE | -15.48 |
| Energy contribution | -15.47 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.800406 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20902779 104 + 22224390 CUC-----UUUUGGCGCUCUUGUCUCUUCAAUCUCUAUUCACAUCAAACACCUCCUGCCAGUUGGCUUAAUUGCCUUUGUCCUUGGCUUUUGCGCUUUUU--------GAUCAGGAU ...-----....((((....))))............................(((((.(((..(((.(((..(((.........)))..))).)))..))--------)..))))). ( -19.50) >DroSec_CAF1 22043 107 + 1 AUCCUCUAUUUAGGCGCUCCUGCCUCUUAAGU--CUAUUCACAUCAUACACCUCCUGCCAGUUGGCUUAAUUGCCUUUGUCCUUGGCUUUUGCGCUUUUU--------GAUCAGGAU ...........(((((....))))).......--..................(((((.(((..(((.(((..(((.........)))..))).)))..))--------)..))))). ( -22.90) >DroSim_CAF1 23925 107 + 1 CUCCUCUAUUUAGGCGCUCCUGCCUCUUAAAU--CUAUUCACAUCAUACACCUCCUGCCAGUUGGCUUAAUUGCCUUUGUCCUUGGCUUUUGCGCUUUUU--------GAUCAGGAU ...........(((((....))))).......--..................(((((.(((..(((.(((..(((.........)))..))).)))..))--------)..))))). ( -22.90) >DroYak_CAF1 19248 111 + 1 CG-UUUCAUUUUGGCGCACUUGCCCUCCAAAU--CAAUGCACAUCAUGCACCACCUGCCAGUUGGCUUCAUGGCCUUUGUCCUUGGCUUUUGCGCUUUUUUUUUUUUUGAUUAG--- ..-.........((((((...(((...((((.--...((((.....))))(((...(((....)))....)))..)))).....)))...))))))..................--- ( -23.00) >consensus CUCCUCUAUUUAGGCGCUCCUGCCUCUUAAAU__CUAUUCACAUCAUACACCUCCUGCCAGUUGGCUUAAUUGCCUUUGUCCUUGGCUUUUGCGCUUUUU________GAUCAGGAU ............(((((.......................................(((....)))......(((.........)))....)))))..................... (-15.48 = -15.47 + -0.00)

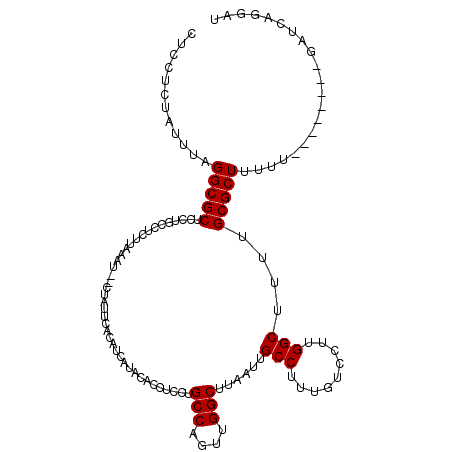

| Location | 20,902,779 – 20,902,883 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 81.40 |
| Mean single sequence MFE | -24.96 |
| Consensus MFE | -18.70 |
| Energy contribution | -19.21 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.814575 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20902779 104 - 22224390 AUCCUGAUC--------AAAAAGCGCAAAAGCCAAGGACAAAGGCAAUUAAGCCAACUGGCAGGAGGUGUUUGAUGUGAAUAGAGAUUGAAGAGACAAGAGCGCCAAAA-----GAG .(((((.((--------(....((......))..........(((......)))...))))))))(((((((..(((.................))).)))))))....-----... ( -23.43) >DroSec_CAF1 22043 107 - 1 AUCCUGAUC--------AAAAAGCGCAAAAGCCAAGGACAAAGGCAAUUAAGCCAACUGGCAGGAGGUGUAUGAUGUGAAUAG--ACUUAAGAGGCAGGAGCGCCUAAAUAGAGGAU (((((.(((--------(....((((....(((.........)))......(((....))).....)))).))))........--.((....((((......))))....))))))) ( -24.60) >DroSim_CAF1 23925 107 - 1 AUCCUGAUC--------AAAAAGCGCAAAAGCCAAGGACAAAGGCAAUUAAGCCAACUGGCAGGAGGUGUAUGAUGUGAAUAG--AUUUAAGAGGCAGGAGCGCCUAAAUAGAGGAG .((((.(((--------(....((((....(((.........)))......(((....))).....)))).))))........--.......((((......))))......)))). ( -23.40) >DroYak_CAF1 19248 111 - 1 ---CUAAUCAAAAAAAAAAAAAGCGCAAAAGCCAAGGACAAAGGCCAUGAAGCCAACUGGCAGGUGGUGCAUGAUGUGCAUUG--AUUUGGAGGGCAAGUGCGCCAAAAUGAAA-CG ---...................(((((...(((.....((((.(((((...(((....)))..)))))((((...))))....--.))))...)))...)))))..........-.. ( -28.40) >consensus AUCCUGAUC________AAAAAGCGCAAAAGCCAAGGACAAAGGCAAUUAAGCCAACUGGCAGGAGGUGUAUGAUGUGAAUAG__AUUUAAGAGGCAAGAGCGCCAAAAUAGAGGAG .((((.........................(((.........)))......(((....)))))))(((((((..(((.................))).)))))))............ (-18.70 = -19.21 + 0.50)

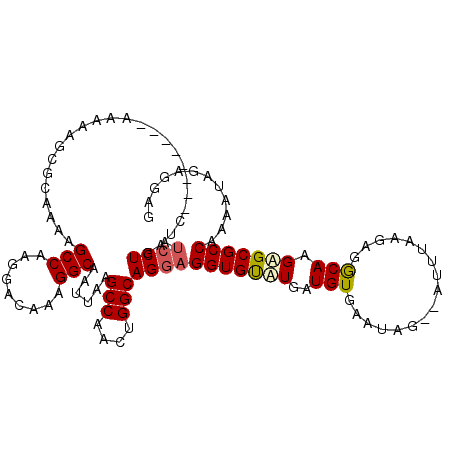
| Location | 20,902,811 – 20,902,921 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 90.30 |
| Mean single sequence MFE | -24.15 |
| Consensus MFE | -19.74 |
| Energy contribution | -20.05 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.595523 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20902811 110 + 22224390 UUCACAUCAAACACCUCCUGCCAGUUGGCUUAAUUGCCUUUGUCCUUGGCUUUUGCGCUUUUUGAUCAGGAUAAGCACUCCGGCAAUAAUCGUCGGCUUUUGUUUUCCAU ...............(((((.(((..(((.(((..(((.........)))..))).)))..)))..))))).(((((..(((((.......)))))....)))))..... ( -25.80) >DroSec_CAF1 22078 110 + 1 UUCACAUCAUACACCUCCUGCCAGUUGGCUUAAUUGCCUUUGUCCUUGGCUUUUGCGCUUUUUGAUCAGGAUAAGCACUCCGGCAAUAAUCGUCGGCUUUUGUUUUCCUU ...............(((((.(((..(((.(((..(((.........)))..))).)))..)))..))))).(((((..(((((.......)))))....)))))..... ( -25.80) >DroSim_CAF1 23960 110 + 1 UUCACAUCAUACACCUCCUGCCAGUUGGCUUAAUUGCCUUUGUCCUUGGCUUUUGCGCUUUUUGAUCAGGAUAAGCACUCCGGCAAUAAUCGUCGGCUUUUGUUUUCCAU ...............(((((.(((..(((.(((..(((.........)))..))).)))..)))..))))).(((((..(((((.......)))))....)))))..... ( -25.80) >DroEre_CAF1 14878 107 + 1 --CACAUCAGACGCCACCUGCCAGUUGGCUUACAUGCCUUUGUUCUUGGCU-UUGCGCUUUUUGAUUAGUUUAAUCACUCUGCCAAUAAUCAGCUGCUUUUGUUUUCCGC --.....((((.((...(((...((((((......(((.........))).-..........((((((...))))))....))))))...)))..)).))))........ ( -19.20) >consensus UUCACAUCAUACACCUCCUGCCAGUUGGCUUAAUUGCCUUUGUCCUUGGCUUUUGCGCUUUUUGAUCAGGAUAAGCACUCCGGCAAUAAUCGUCGGCUUUUGUUUUCCAU ...............(((((.(((..(((.(((..(((.........)))..))).)))..)))..))))).(((((..(((((.......)))))....)))))..... (-19.74 = -20.05 + 0.31)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:03:36 2006