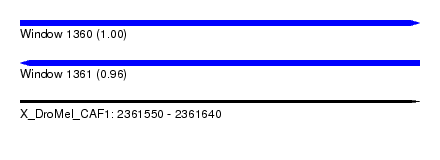
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 2,361,550 – 2,361,640 |
| Length | 90 |
| Max. P | 0.995855 |

| Location | 2,361,550 – 2,361,640 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 72.13 |
| Mean single sequence MFE | -25.02 |
| Consensus MFE | -12.96 |
| Energy contribution | -13.88 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.52 |
| SVM decision value | 2.62 |
| SVM RNA-class probability | 0.995855 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
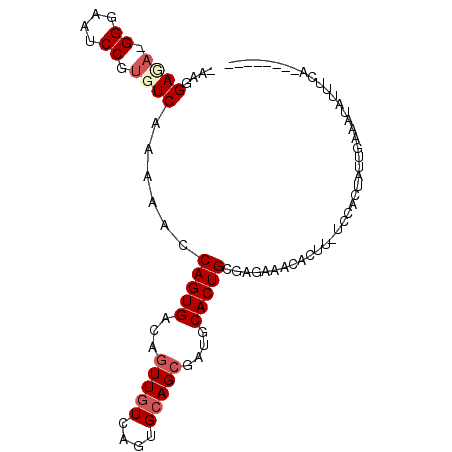
>X_DroMel_CAF1 2361550 90 + 22224390 -AAGGAUA-GGGAAUCCGUGUCAAAAACCAGUGACAGUUGUCAGUGCAGCGAUGCACUGCGAGAAACACUUGUAUUGUAUUCAUGUUAUUUA-------- -...((((-.((...)).)))).......((((((((((((....)))))((((((.((((((.....)))))).))))))..)))))))..-------- ( -23.60) >DroSec_CAF1 6133 89 + 1 -AAGGAUA-GGGAAUCCGUGUCAAAAACCAGUGACAGUUGUCAGUGCAGCGAUGCACUGCGAGAAACACUU-UCCUCUGUUGAAGUAUUUCA-------- -..((((.-....)))).(((((........))))).(((.(((((((....))))))))))((((.((((-(........))))).)))).-------- ( -25.20) >DroEre_CAF1 6849 96 + 1 -AAGGAGA-GGGAAUCCGUGUCAAAAACCAGUGACAGUUGUCAGUGCAGCGAUGCACUGGCAGAAACACUU-UCUACUCUUGAAAAAUUUCAU-UGCCUG -.((((((-(((((....(((((........))))).(((((((((((....))))))))))).......)-))).))))((((....)))).-..))). ( -29.10) >DroYak_CAF1 6497 98 + 1 GAAGGAGC-GGGAAUCCGUGUCAAAAACCAGUGACAGUUGUCAGUGCAGCGAUACACUGCAAGAAACACUU-UCCAGUAUCGAAAUAUGUCAUAUGCCUG ..(((.((-((....))))...........((((((....((..(((((.......))))).((.((....-....)).))))....))))))...))). ( -22.00) >DroAna_CAF1 5619 71 + 1 GAAAGACAGGGGAAACCGUGUCAAAAACCAGUGACAGUUGUCAGUGAAG----ACACUGAGAGAAAAGU-----------------GGGGGG-------- ....((((.((....)).)))).....(((.(.....((.((((((...----.)))))).))...).)-----------------))....-------- ( -25.20) >consensus _AAGGAGA_GGGAAUCCGUGUCAAAAACCAGUGACAGUUGUCAGUGCAGCGAUGCACUGCGAGAAACACUU_UCCACUAUUGAAAUAUUUCA________ ....((((.((....)).))))......(((((...(((((....)))))....)))))......................................... (-12.96 = -13.88 + 0.92)

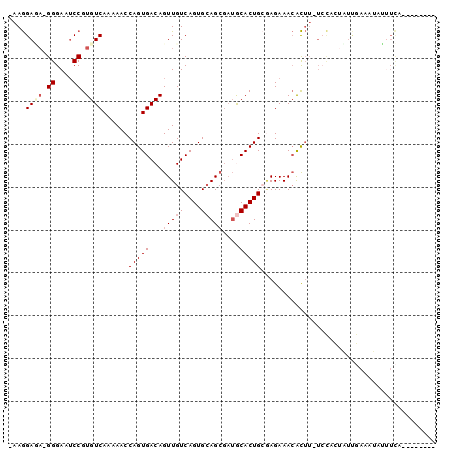
| Location | 2,361,550 – 2,361,640 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 72.13 |
| Mean single sequence MFE | -21.31 |
| Consensus MFE | -12.98 |
| Energy contribution | -13.62 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.959733 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
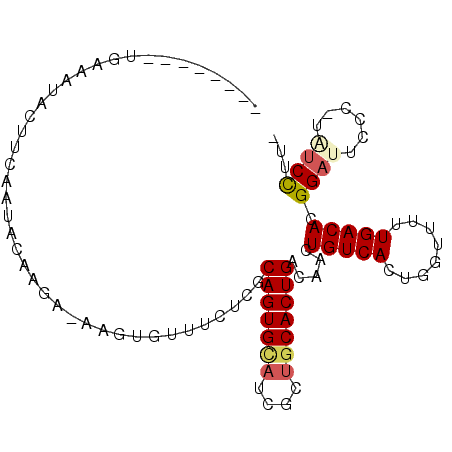
>X_DroMel_CAF1 2361550 90 - 22224390 --------UAAAUAACAUGAAUACAAUACAAGUGUUUCUCGCAGUGCAUCGCUGCACUGACAACUGUCACUGGUUUUUGACACGGAUUCCC-UAUCCUU- --------...............(((.((.((((.......(((((((....)))))))........)))).))..)))....((((....-.))))..- ( -19.96) >DroSec_CAF1 6133 89 - 1 --------UGAAAUACUUCAACAGAGGA-AAGUGUUUCUCGCAGUGCAUCGCUGCACUGACAACUGUCACUGGUUUUUGACACGGAUUCCC-UAUCCUU- --------.((((((((((.......).-)))))))))..((((((((....))))))).)...(((((........))))).((((....-.))))..- ( -24.40) >DroEre_CAF1 6849 96 - 1 CAGGCA-AUGAAAUUUUUCAAGAGUAGA-AAGUGUUUCUGCCAGUGCAUCGCUGCACUGACAACUGUCACUGGUUUUUGACACGGAUUCCC-UCUCCUU- .(((..-.((((....))))((((..((-..(((((...(((((((((....)))))((((....)))).))))....)))))...))..)-)))))).- ( -25.10) >DroYak_CAF1 6497 98 - 1 CAGGCAUAUGACAUAUUUCGAUACUGGA-AAGUGUUUCUUGCAGUGUAUCGCUGCACUGACAACUGUCACUGGUUUUUGACACGGAUUCCC-GCUCCUUC ..(((...(((((......((((((...-.))))))..((((((((((....))))))).))).)))))((((((...))).)))......-)))..... ( -24.00) >DroAna_CAF1 5619 71 - 1 --------CCCCCC-----------------ACUUUUCUCUCAGUGU----CUUCACUGACAACUGUCACUGGUUUUUGACACGGUUUCCCCUGUCUUUC --------....((-----------------(........((((((.----...))))))..........))).....((((.((....)).)))).... ( -13.07) >consensus ________UGAAAUACUUCAAUACAAGA_AAGUGUUUCUCGCAGUGCAUCGCUGCACUGACAACUGUCACUGGUUUUUGACACGGAUUCCC_UAUCCUU_ .........................................(((((((....))))))).....(((((........))))).((((......))))... (-12.98 = -13.62 + 0.64)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:55:07 2006