
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,902,230 – 20,902,391 |
| Length | 161 |
| Max. P | 0.896537 |

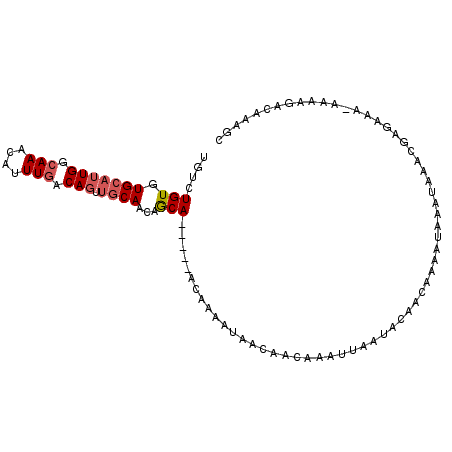
| Location | 20,902,230 – 20,902,329 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 81.06 |
| Mean single sequence MFE | -14.60 |
| Consensus MFE | -4.76 |
| Energy contribution | -5.40 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.33 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.736946 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
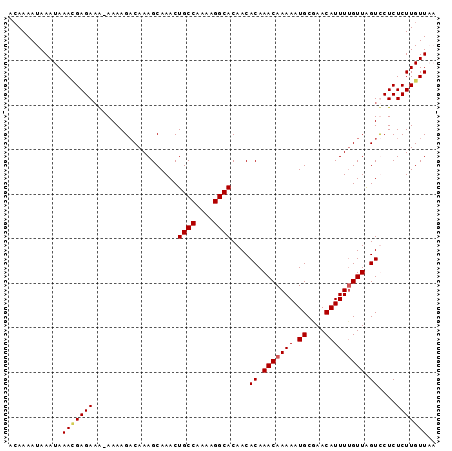
>X_DroMel_CAF1 20902230 99 + 22224390 UGUCUGUGUGCAUUGGCAAACAUUUGACAGUUGCAACAGCA-----ACAAAAUAACAACAAAUUAAUGCAACAAAAUAAAUAAAAGAGAAAAAAAAGACAAAGC (((((.(((((((((((((....)))...(((((....)))-----))..............))))))).)))......................))))).... ( -16.20) >DroSec_CAF1 21472 98 + 1 UGUCUGUGUGCAUUGGCAAACAUUUAACAGUUGCAACAGCA-----ACAAAAUAACAACAAAUUAAUACAACAAAAUAAAUAAACGAGAAA-AAAAGACAAAGC (((((((.(((....))).))........(((((....)))-----))...........................................-...))))).... ( -14.90) >DroSim_CAF1 23354 98 + 1 UGUCUGUGUGCAUUGGCAAACAUUUGACAGUUGCAACAGCA-----ACAAAAUAACAACAAAUUAAUACAACAAAAUAAAUAAACGAGAAA-AAAAGACAAAGC (((((((.(((....))).))........(((((....)))-----))...........................................-...))))).... ( -14.90) >DroEre_CAF1 14393 97 + 1 UGUCUGUGUGCAUUGGCAAACAUUUGACAGUUGCAACAGCA-----ACAAAAUAACAACAAAUUAAUACAAAAAAAUAAAUAAAUGAGA-G-AAAAGCCAAAGC .........((.(((((............(((((....)))-----))....................................(....-)-....))))).)) ( -14.00) >DroAna_CAF1 12665 85 + 1 UCGGUGUGUGGAUUGGCAAGCAUUUGACA----CAAAAACACACACAAAAAAUAACAAAAAGUUGAC----UAAAAUAA---AAUGGGC-U-A------AAAGU ...(((((((..(((.(((....)))...----)))...)))))))......((((.....))))..----........---.......-.-.------..... ( -13.00) >consensus UGUCUGUGUGCAUUGGCAAACAUUUGACAGUUGCAACAGCA_____ACAAAAUAACAACAAAUUAAUACAACAAAAUAAAUAAACGAGAAA_AAAAGACAAAGC ....(((.(((((((.(((....))).))).))))...)))............................................................... ( -4.76 = -5.40 + 0.64)


| Location | 20,902,295 – 20,902,391 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 92.67 |
| Mean single sequence MFE | -15.78 |
| Consensus MFE | -12.05 |
| Energy contribution | -12.42 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.28 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.856187 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20902295 96 + 22224390 ACAAAAUAAAUAAAAGAGAAAAAAAAGACAAAGCAAACUGCCAAAAGGCACAACACAAACAAAAAUGCAAACAUUUUGUUAGUCCUCUCUUGUUAA ........(((((.((((....................((((....))))....((.(((((((.((....))))))))).)).)))).))))).. ( -15.00) >DroSec_CAF1 21537 95 + 1 ACAAAAUAAAUAAACGAGAAA-AAAAGACAAAGCAAACUGCCAAAAGGCACAACACAAACAAAAAUGCGAACAUUUGGUUAGUUCUCUCUUGUUAA ............(((((((..-................((((....))))....((.(((..(((((....))))).))).))....))))))).. ( -13.50) >DroSim_CAF1 23419 95 + 1 ACAAAAUAAAUAAACGAGAAA-AAAAGACAAAGCAAACUGCCAAAAGGCACAACACAAACAAAAAUGCGAACAUUUUGUUAGUUCUCUCUUGUUCC ............(((((((..-................((((....))))....((.(((((((.((....))))))))).))....))))))).. ( -14.60) >DroEre_CAF1 14458 94 + 1 AAAAAAUAAAUAAAUGAGA-G-AAAAGCCAAAGCAAACUGCCAAAAGGCACAACACAAACAAAAAUGCGAACAUUUUGUUGGUCCUCUCUUGUUCU ............((..(((-(-(...(((((.......((((....))))...........((((((....)))))).)))))..)))))..)).. ( -20.00) >consensus ACAAAAUAAAUAAACGAGAAA_AAAAGACAAAGCAAACUGCCAAAAGGCACAACACAAACAAAAAUGCGAACAUUUUGUUAGUCCUCUCUUGUUAA ............(((((((...................((((....))))....((.(((((((.((....))))))))).))....))))))).. (-12.05 = -12.42 + 0.37)


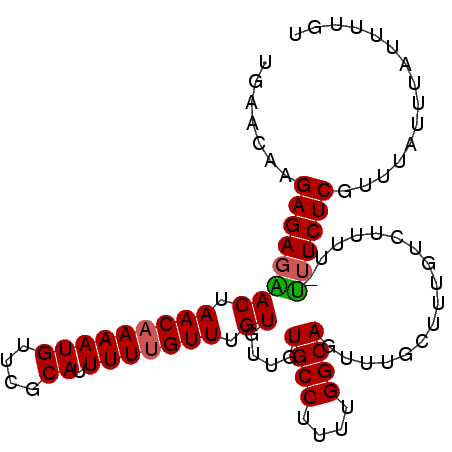
| Location | 20,902,295 – 20,902,391 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 92.67 |
| Mean single sequence MFE | -19.05 |
| Consensus MFE | -16.29 |
| Energy contribution | -16.35 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.896537 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
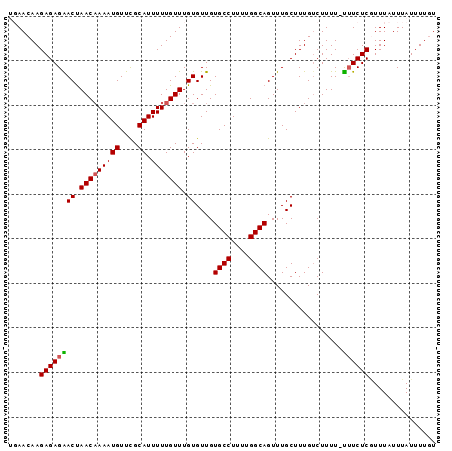
>X_DroMel_CAF1 20902295 96 - 22224390 UUAACAAGAGAGGACUAACAAAAUGUUUGCAUUUUUGUUUGUGUUGUGCCUUUUGGCAGUUUGCUUUGUCUUUUUUUUCUCUUUUAUUUAUUUUGU .....((((((((((.(((((((((....)).))))))).))....((((....))))...................))))))))........... ( -18.60) >DroSec_CAF1 21537 95 - 1 UUAACAAGAGAGAACUAACCAAAUGUUCGCAUUUUUGUUUGUGUUGUGCCUUUUGGCAGUUUGCUUUGUCUUUU-UUUCUCGUUUAUUUAUUUUGU .......(((((((..(..((((.((.((((........))))...((((....))))....))))))..)..)-))))))............... ( -17.10) >DroSim_CAF1 23419 95 - 1 GGAACAAGAGAGAACUAACAAAAUGUUCGCAUUUUUGUUUGUGUUGUGCCUUUUGGCAGUUUGCUUUGUCUUUU-UUUCUCGUUUAUUUAUUUUGU .((((.(((((((((.(((((((((....)).))))))).))....((((....))))..............))-))))).))))........... ( -19.30) >DroEre_CAF1 14458 94 - 1 AGAACAAGAGAGGACCAACAAAAUGUUCGCAUUUUUGUUUGUGUUGUGCCUUUUGGCAGUUUGCUUUGGCUUUU-C-UCUCAUUUAUUUAUUUUUU .......((((((((.(((((((((....)).))))))).)).....(((....(((.....)))..)))...)-)-))))............... ( -21.20) >consensus UGAACAAGAGAGAACUAACAAAAUGUUCGCAUUUUUGUUUGUGUUGUGCCUUUUGGCAGUUUGCUUUGUCUUUU_UUUCUCGUUUAUUUAUUUUGU .......((((((((.(((((((((....)).))))))).))....((((....)))).................))))))............... (-16.29 = -16.35 + 0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:03:34 2006