
| Sequence ID | X_DroMel_CAF1 |
|---|---|
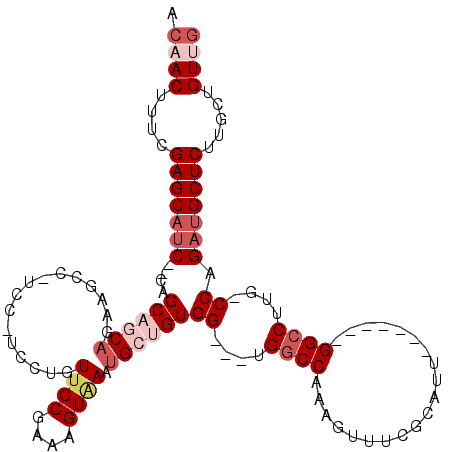
| Location | 20,901,429 – 20,901,536 |
| Length | 107 |
| Max. P | 0.933158 |

| Location | 20,901,429 – 20,901,536 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 81.17 |
| Mean single sequence MFE | -36.10 |
| Consensus MFE | -20.62 |
| Energy contribution | -22.86 |
| Covariance contribution | 2.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.647263 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20901429 107 + 22224390 ---ACUUUCGAGCAUCAGCAGCACCAGAAGCCAUCCAUCCUGUUGCGAAAGUGAAUGCUGUGGUGGUGGCCAAAGUUUCGCAUU-------GGCCUUG-CCAGAUGCUCUUGCUGUUG ---......(((((((.((((((.(((............)))(..(....)..).))))))((..(.((((((.(.....).))-------)))))..-)).)))))))......... ( -40.30) >DroSec_CAF1 20719 101 + 1 ACAACUUUCGAGCAUC--CAGCCGCAGAAGCCGUC----CUGUUGCGAAAGUUAAUGCUGUGG---UGGCCAAAGUUUCGCAUU-------GGCCUUG-CCAGAUGCUCUUGCUGUUG .((((....(((((((--((((.((((........----)))).((....))....))))(((---(((((((.(.....).))-------))))..)-)))))))))).....)))) ( -36.10) >DroSim_CAF1 22601 107 + 1 ACAACUUUCGAGCAUC--CAGCAGCAGAAGCCGUC-----UGUUGCGAAAGUGAAUGCUGUGG---UGGCCAAAGUUUCGCAAUGGCCAAUGGCCUUG-CCAGAUGCUCGUGCUGUUG .((((...((((((((--..((((((((.....))-----))))))((((.(....(((....---.)))...).))))((((.((((...)))))))-)..))))))))....)))) ( -43.10) >DroEre_CAF1 13580 96 + 1 ACAACUUU-GAGCCUC---AGCCUCUG--------CCUCUUGUUGCGAAAGUAAAUGCUGUGG---UGGCCAAAGUUUCGCCUU-------GGCCUUGUCCAGAUGCUCCUGCUGUUG .((((...-((((.((---.(((.(.(--------(......((((....))))..)).).))---)((((((.(.....).))-------)))).......)).)))).....)))) ( -29.00) >DroYak_CAF1 17974 101 + 1 ACAACUUUCGAGCAUC---AGCAGCAGAAGCC---CAUCUUGUUGCGAAAGUAAAUGCUGUGG---UGCCCAAAGUUUCGCCUU-------GGCCUUG-CCAGCUGCUCUUGCUGUAG ..........((((..---((((((....(((---(((....((((....)))).....))))---.))..............(-------(((...)-)))))))))..)))).... ( -32.00) >consensus ACAACUUUCGAGCAUC__CAGCAGCAGAAGCC_UCC_UCCUGUUGCGAAAGUAAAUGCUGUGG___UGGCCAAAGUUUCGCAUU_______GGCCUUG_CCAGAUGCUCUUGCUGUUG .((((....(((((((....((((((................((((....)))).))))))((....((((....................))))....)).))))))).....)))) (-20.62 = -22.86 + 2.24)


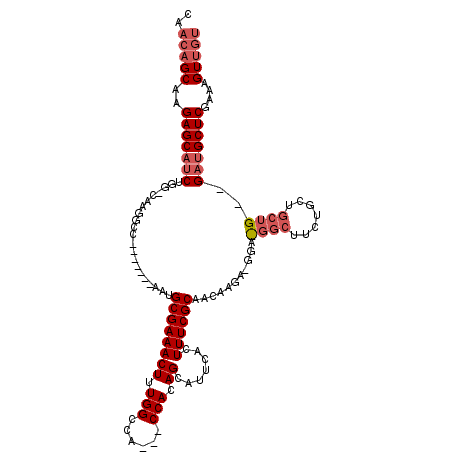
| Location | 20,901,429 – 20,901,536 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 81.17 |
| Mean single sequence MFE | -36.36 |
| Consensus MFE | -19.35 |
| Energy contribution | -20.97 |
| Covariance contribution | 1.62 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.53 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.933158 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20901429 107 - 22224390 CAACAGCAAGAGCAUCUGG-CAAGGCC-------AAUGCGAAACUUUGGCCACCACCACAGCAUUCACUUUCGCAACAGGAUGGAUGGCUUCUGGUGCUGCUGAUGCUCGAAAGU--- .........(((((((.((-((.((((-------((.........))))))..(((((.((((((((..(((......)))))))).)))..))))).)))))))))))......--- ( -38.60) >DroSec_CAF1 20719 101 - 1 CAACAGCAAGAGCAUCUGG-CAAGGCC-------AAUGCGAAACUUUGGCCA---CCACAGCAUUAACUUUCGCAACAG----GACGGCUUCUGCGGCUG--GAUGCUCGAAAGUUGU ..(((((..(((((((..(-(..((((-------((.........)))))).---................((((..((----.....))..))))))..--)))))))....))))) ( -37.70) >DroSim_CAF1 22601 107 - 1 CAACAGCACGAGCAUCUGG-CAAGGCCAUUGGCCAUUGCGAAACUUUGGCCA---CCACAGCAUUCACUUUCGCAACA-----GACGGCUUCUGCUGCUG--GAUGCUCGAAAGUUGU ..(((((.((((((((..(-((.((((...)))).((((((((.....((..---.....))......))))))))((-----((.....)))).)))..--))))))))...))))) ( -42.90) >DroEre_CAF1 13580 96 - 1 CAACAGCAGGAGCAUCUGGACAAGGCC-------AAGGCGAAACUUUGGCCA---CCACAGCAUUUACUUUCGCAACAAGAGG--------CAGAGGCU---GAGGCUC-AAAGUUGU ..(((((..((((...(((....((((-------((((.....)))))))).---)))((((.(((.((((........))))--------.))).)))---)..))))-...))))) ( -31.90) >DroYak_CAF1 17974 101 - 1 CUACAGCAAGAGCAGCUGG-CAAGGCC-------AAGGCGAAACUUUGGGCA---CCACAGCAUUUACUUUCGCAACAAGAUG---GGCUUCUGCUGCU---GAUGCUCGAAAGUUGU ..(((((..(((((((((.-...(.((-------((((.....)))))).).---...))))..........(((...(((..---....)))..))).---..)))))....))))) ( -30.70) >consensus CAACAGCAAGAGCAUCUGG_CAAGGCC_______AAUGCGAAACUUUGGCCA___CCACAGCAUUCACUUUCGCAACAAGA_GGA_GGCUUCUGCUGCUG__GAUGCUCGAAAGUUGU ..(((((..(((((((.....................((((((((.(((......))).)).......))))))...........((((.......))))..)))))))....))))) (-19.35 = -20.97 + 1.62)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:03:30 2006