
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,884,131 – 20,884,324 |
| Length | 193 |
| Max. P | 0.951906 |

| Location | 20,884,131 – 20,884,244 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.27 |
| Mean single sequence MFE | -40.38 |
| Consensus MFE | -31.65 |
| Energy contribution | -33.65 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.751985 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20884131 113 + 22224390 AAUCCGCGGCAAUGUCCGACG-------ACCGACUGGCGACCGGCGACUGUUGCAAAUCGAAACGAUUCGCUGGAUCCGCUGGAUUCGCCAGCAAAACCUGCCCCGGAUCACAAUUUGUC .(((((.((((..(((....)-------)).(.(((((((((((((......((.(((((...))))).))......))))))..))))))))......)))).)))))........... ( -41.90) >DroSec_CAF1 4185 120 + 1 AAUCCGCGGCAAUGUCCGACGACCGACGACCGACUGGCGACCGGCGACUGUUGCAAAUCGAAACGAUUCGCUGGAUCCGCUGGAUUCGCCAGCAAAACCUGCCCCGGAUCACAAUUUGUC .(((((.((((.((((........))))...(.(((((((((((((......((.(((((...))))).))......))))))..))))))))......)))).)))))........... ( -43.10) >DroSim_CAF1 4787 113 + 1 AAUCCGCGGCAAUGUCCGACG-------ACCGACUGGCGACCGGCGACUGUUGCAAAUCGAAACGAUUCGCUGGAUCCGCUGGAUUCGCCAGCAAAACCUGCCCCGGAUCACAAUUUGUC .(((((.((((..(((....)-------)).(.(((((((((((((......((.(((((...))))).))......))))))..))))))))......)))).)))))........... ( -41.90) >DroYak_CAF1 5019 104 + 1 AAUCCGCGGCAAUGUCCGAUG-------ACCGACUGGCGACCGGCGACUGUUGCAAAUCGAAACGAUUCGCUGGAUCC---------GCCAGCAAAACCUGCACCGGAUCACAAUUUGUC .(((((..(((..(((....)-------)).(.((((((.(((((((.((((.........))))..)))))))...)---------))))))......)))..)))))........... ( -34.60) >consensus AAUCCGCGGCAAUGUCCGACG_______ACCGACUGGCGACCGGCGACUGUUGCAAAUCGAAACGAUUCGCUGGAUCCGCUGGAUUCGCCAGCAAAACCUGCCCCGGAUCACAAUUUGUC .(((((.((((.....((............)).(((((((((((((......((.(((((...))))).))......))))))..))))))).......)))).)))))........... (-31.65 = -33.65 + 2.00)

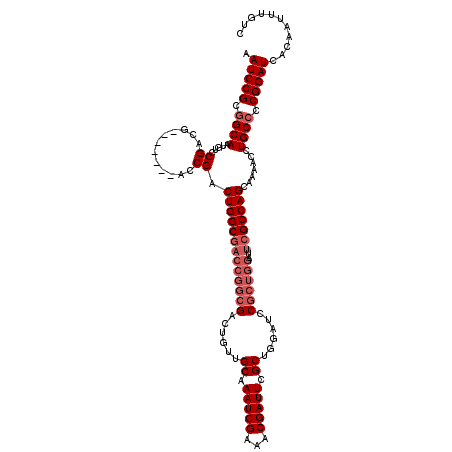

| Location | 20,884,131 – 20,884,244 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.27 |
| Mean single sequence MFE | -47.55 |
| Consensus MFE | -41.00 |
| Energy contribution | -41.75 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.951906 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
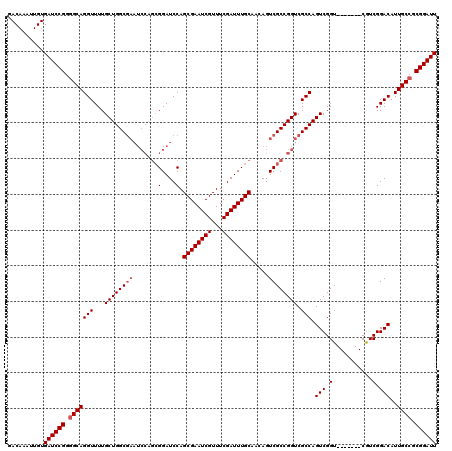
>X_DroMel_CAF1 20884131 113 - 22224390 GACAAAUUGUGAUCCGGGGCAGGUUUUGCUGGCGAAUCCAGCGGAUCCAGCGAAUCGUUUCGAUUUGCAACAGUCGCCGGUCGCCAGUCGGU-------CGUCGGACAUUGCCGCGGAUU ..........((((((.(((((.....((((((((..((.((((.....((((((((...)))))))).....)))).))))))))))..((-------(....))).))))).)))))) ( -49.60) >DroSec_CAF1 4185 120 - 1 GACAAAUUGUGAUCCGGGGCAGGUUUUGCUGGCGAAUCCAGCGGAUCCAGCGAAUCGUUUCGAUUUGCAACAGUCGCCGGUCGCCAGUCGGUCGUCGGUCGUCGGACAUUGCCGCGGAUU ..........((((((.(((((..((((((((.....))))))))(((.((((((((...))))))))....(.((((((..(((....)))..)))).)).))))..))))).)))))) ( -51.30) >DroSim_CAF1 4787 113 - 1 GACAAAUUGUGAUCCGGGGCAGGUUUUGCUGGCGAAUCCAGCGGAUCCAGCGAAUCGUUUCGAUUUGCAACAGUCGCCGGUCGCCAGUCGGU-------CGUCGGACAUUGCCGCGGAUU ..........((((((.(((((.....((((((((..((.((((.....((((((((...)))))))).....)))).))))))))))..((-------(....))).))))).)))))) ( -49.60) >DroYak_CAF1 5019 104 - 1 GACAAAUUGUGAUCCGGUGCAGGUUUUGCUGGC---------GGAUCCAGCGAAUCGUUUCGAUUUGCAACAGUCGCCGGUCGCCAGUCGGU-------CAUCGGACAUUGCCGCGGAUU ..........(((((((.((((((...((((((---------((.....((((((((...)))))))).....)))))))).))).(((.(.-------...).)))..)))).)))))) ( -39.70) >consensus GACAAAUUGUGAUCCGGGGCAGGUUUUGCUGGCGAAUCCAGCGGAUCCAGCGAAUCGUUUCGAUUUGCAACAGUCGCCGGUCGCCAGUCGGU_______CGUCGGACAUUGCCGCGGAUU ..........((((((.(((((((...((((((((.......(....).((((((((...)))))))).....)))))))).))).(((.(...........).)))..)))).)))))) (-41.00 = -41.75 + 0.75)


| Location | 20,884,164 – 20,884,284 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.00 |
| Mean single sequence MFE | -41.35 |
| Consensus MFE | -35.59 |
| Energy contribution | -36.40 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.886029 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20884164 120 - 22224390 AUAAAUAAAUAACAUAAUCAAGAGGCUUGCCCGGGCGUCGGACAAAUUGUGAUCCGGGGCAGGUUUUGCUGGCGAAUCCAGCGGAUCCAGCGAAUCGUUUCGAUUUGCAACAGUCGCCGG .....................(((((((((((.((.((((.(.....).)))))).))))))))))).(((((((((((...))))...((((((((...)))))))).....))))))) ( -44.00) >DroSec_CAF1 4225 120 - 1 AUAAAUAAAUAACAUAAUCAAGAGGCUUGCCCGGGCGUCGGACAAAUUGUGAUCCGGGGCAGGUUUUGCUGGCGAAUCCAGCGGAUCCAGCGAAUCGUUUCGAUUUGCAACAGUCGCCGG .....................(((((((((((.((.((((.(.....).)))))).))))))))))).(((((((((((...))))...((((((((...)))))))).....))))))) ( -44.00) >DroSim_CAF1 4820 120 - 1 AUAAAUAAAUAACAUAAUCAAGAGGCUUGCCCGGGCGUCGGACAAAUUGUGAUCCGGGGCAGGUUUUGCUGGCGAAUCCAGCGGAUCCAGCGAAUCGUUUCGAUUUGCAACAGUCGCCGG .....................(((((((((((.((.((((.(.....).)))))).))))))))))).(((((((((((...))))...((((((((...)))))))).....))))))) ( -44.00) >DroYak_CAF1 5052 111 - 1 AUAAAUAAAUAACAUAAUCAAGAGGUUUCCCCGGGCGUCGGACAAAUUGUGAUCCGGUGCAGGUUUUGCUGGC---------GGAUCCAGCGAAUCGUUUCGAUUUGCAACAGUCGCCGG ............((.((((....((.....))..((..((((((.....)).))))..)).)))).))(((((---------((.....((((((((...)))))))).....))))))) ( -33.40) >consensus AUAAAUAAAUAACAUAAUCAAGAGGCUUGCCCGGGCGUCGGACAAAUUGUGAUCCGGGGCAGGUUUUGCUGGCGAAUCCAGCGGAUCCAGCGAAUCGUUUCGAUUUGCAACAGUCGCCGG .....................(((((((((((.((.((((.(.....).)))))).))))))))))).(((((((.......(....).((((((((...)))))))).....))))))) (-35.59 = -36.40 + 0.81)

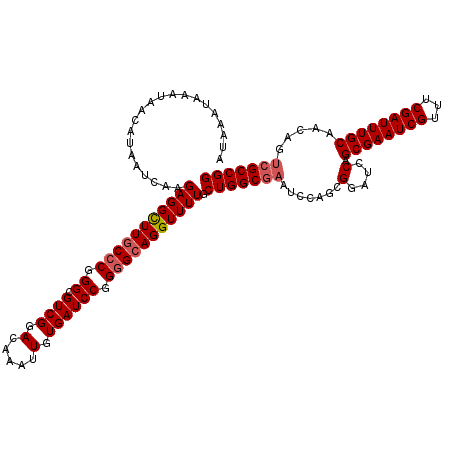
| Location | 20,884,204 – 20,884,324 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.00 |
| Mean single sequence MFE | -32.34 |
| Consensus MFE | -29.31 |
| Energy contribution | -29.68 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.648831 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
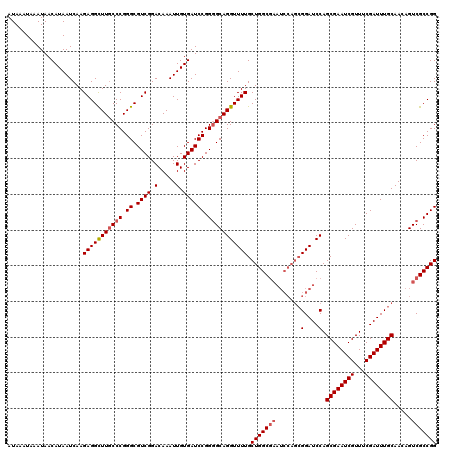
>X_DroMel_CAF1 20884204 120 - 22224390 ACGCGCGGCAAUAACAUUAAACAUUAACGAGCAUGGCUAAAUAAAUAAAUAACAUAAUCAAGAGGCUUGCCCGGGCGUCGGACAAAUUGUGAUCCGGGGCAGGUUUUGCUGGCGAAUCCA .(((.((((...................((..(((.................)))..))..(((((((((((.((.((((.(.....).)))))).))))))))))))))))))...... ( -35.13) >DroSec_CAF1 4265 120 - 1 ACGCGCGGCAAUAACAUUAAACAUUAACGAGCAUGUCUAAAUAAAUAAAUAACAUAAUCAAGAGGCUUGCCCGGGCGUCGGACAAAUUGUGAUCCGGGGCAGGUUUUGCUGGCGAAUCCA .(((.((((...................((..((((...............))))..))..(((((((((((.((.((((.(.....).)))))).))))))))))))))))))...... ( -35.26) >DroSim_CAF1 4860 120 - 1 ACGCGCGGCAAUAACAUUAAACAUUAACGAGCAUGGCUAAAUAAAUAAAUAACAUAAUCAAGAGGCUUGCCCGGGCGUCGGACAAAUUGUGAUCCGGGGCAGGUUUUGCUGGCGAAUCCA .(((.((((...................((..(((.................)))..))..(((((((((((.((.((((.(.....).)))))).))))))))))))))))))...... ( -35.13) >DroYak_CAF1 5090 113 - 1 ACGCGCAGCAAUAACAUUAAACAUUAACGAGCAUGGCUAAAUAAAUAAAUAACAUAAUCAAGAGGUUUCCCCGGGCGUCGGACAAAUUGUGAUCCGGUGCAGGUUUUGCUGGC------- ..((.((((((.................((..(((.................)))..))...........((..((..((((((.....)).))))..)).))..))))))))------- ( -23.83) >consensus ACGCGCGGCAAUAACAUUAAACAUUAACGAGCAUGGCUAAAUAAAUAAAUAACAUAAUCAAGAGGCUUGCCCGGGCGUCGGACAAAUUGUGAUCCGGGGCAGGUUUUGCUGGCGAAUCCA .(((.((((...................((..(((.................)))..))..(((((((((((.((.((((.(.....).)))))).))))))))))))))))))...... (-29.31 = -29.68 + 0.37)

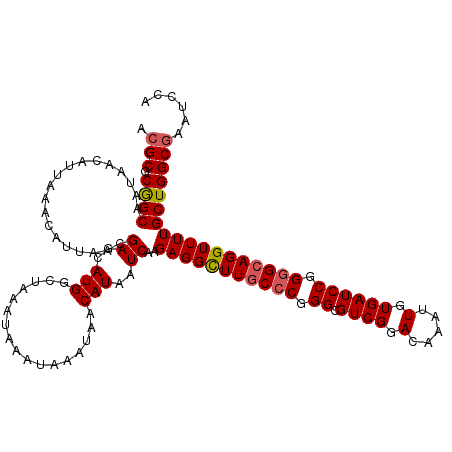
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:03:24 2006