
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,883,219 – 20,883,494 |
| Length | 275 |
| Max. P | 0.788758 |

| Location | 20,883,219 – 20,883,339 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.96 |
| Mean single sequence MFE | -26.58 |
| Consensus MFE | -12.99 |
| Energy contribution | -17.43 |
| Covariance contribution | 4.44 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.49 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.788758 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20883219 120 + 22224390 UCCGGUAAACGGAAUCGUUGGAUUAUUCCUAUUGAUAAGUGAUCAAUUCAUUCAUUUAACUAACUUUCAAUUCAUCCUUAACCGUGUAAACUCAUUCAGGGCUUCAGGCCCAGUGAAUCC ..((((.((.(((.(((..(((....)))...)))(((((((.........)))))))................))))).)))).......(((((..((((.....))))))))).... ( -25.60) >DroSec_CAF1 3318 104 + 1 UCCGGUAAACGAAAACGUUGGAUUGAUCCUUUUGUUAAGUGAUCAAUUGA----------------UCAAUUCAUCUUUUACCGAAUAAACUCAUUCAGGGCUUCAGGUCCAGUGAAUCC ..(((((((.((....(((((((((((((((.....))).))))))))..----------------.))))...)).))))))).......(((((..((((.....))))))))).... ( -26.40) >DroSim_CAF1 3910 104 + 1 UCCGGUAAACGAAAACGUUGGAUUGAUCCUUUUGUUAAGUGAUCAAUUGA----------------UCAAUUCAUCUUUUACCGGAUAAACUUAUUCAGGGCUUCAGGCCCAGUGAAUCC (((((((((.((....(((((((((((((((.....))).))))))))..----------------.))))...)).))))))))).......(((((((((.....))))..))))).. ( -32.90) >DroYak_CAF1 4095 90 + 1 UUCGCUAAACGGAAACGCUGGC-------------CAAGUUAACAGUCCU----------------GCAAUGCAUCUUUAACCGGAUGAACUCAUUCAUGGCUUCAGGCCCAG-GAAACC ...((((..((....)).))))-------------...........((((----------------(................(((((....)))))..(((.....))))))-)).... ( -21.40) >consensus UCCGGUAAACGAAAACGUUGGAUUGAUCCUUUUGUUAAGUGAUCAAUUCA________________UCAAUUCAUCUUUAACCGGAUAAACUCAUUCAGGGCUUCAGGCCCAGUGAAUCC ((((((.((((....))))((((((((((((.....))).)))))))))...............................)))))).....(((((..((((.....))))))))).... (-12.99 = -17.43 + 4.44)

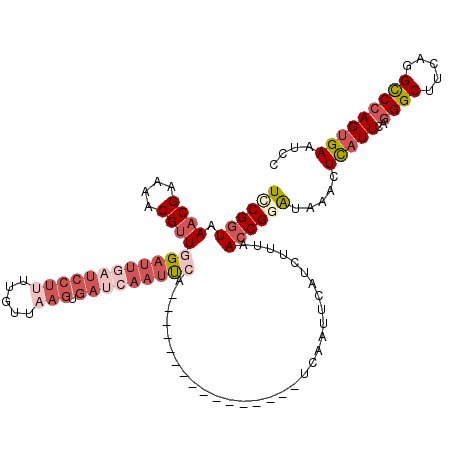
| Location | 20,883,299 – 20,883,416 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.84 |
| Mean single sequence MFE | -34.85 |
| Consensus MFE | -24.83 |
| Energy contribution | -25.57 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.700727 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20883299 117 + 22224390 ACCGUGUAAACUCAUUCAGGGCUUCAGGCCCAGUGAAUCCUUUUUGGCCAUUAAGAAGUCAAUUGAAUCUGCCUCGGGAAGCAACUGUUCCGAUCUCAUUCAGAGAUUGGCCAAAGU--- ...........(((((..((((.....))))))))).....(((((((((...(((..((....)).)))...((((((........))))))((((.....)))).))))))))).--- ( -31.50) >DroSec_CAF1 3382 113 + 1 ACCGAAUAAACUCAUUCAGGGCUUCAGGUCCAGUGAAUCCUUUUUGGCCAUUA----GUCAAUUGAAUCUACCUCGGGAGGCAACUGUUCCCAUCUCAUUCAGAGAUUGGCCAAAGU--- ...........(((((..((((.....))))))))).....(((((((((...----..................(((((....)...))))(((((.....)))))))))))))).--- ( -30.10) >DroSim_CAF1 3974 113 + 1 ACCGGAUAAACUUAUUCAGGGCUUCAGGCCCAGUGAAUCCUUUUUGGCCAUUA----GUCAAUUGAAUCUGCCUCGGGAGGCAACUGUUCCGAUCUCAUUCAGAGAUUGGCCAAAGU--- ...((((..(((......((((.....)))))))..)))).((((((((....----.......((((.(((((....)))))...)))).((((((.....)))))))))))))).--- ( -37.70) >DroYak_CAF1 4146 115 + 1 ACCGGAUGAACUCAUUCAUGGCUUCAGGCCCAG-GAAACCUUUUUGGCCAUUA----GUCAGUUGGAUCUGCCUCGAGAAGAAGCUGAUCCGAUCGCAUUCAGAGAUUGGCCAAAGUGGA .(((((((....)))))..(((.....)))..)-)...((.(((((((((...----(((..(((((((.((.((.....)).)).)))))))((.......)))))))))))))).)). ( -40.10) >consensus ACCGGAUAAACUCAUUCAGGGCUUCAGGCCCAGUGAAUCCUUUUUGGCCAUUA____GUCAAUUGAAUCUGCCUCGGGAAGCAACUGUUCCGAUCUCAUUCAGAGAUUGGCCAAAGU___ ...(((((....))))).((((.....))))..........((((((((..........((.(((..(((......)))..))).))....((((((.....)))))))))))))).... (-24.83 = -25.57 + 0.75)

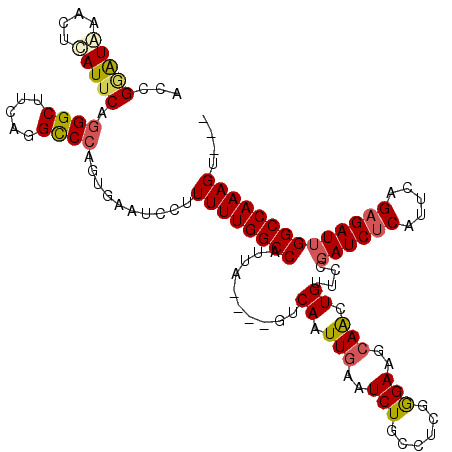
| Location | 20,883,299 – 20,883,416 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.84 |
| Mean single sequence MFE | -34.83 |
| Consensus MFE | -27.89 |
| Energy contribution | -28.70 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.754820 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20883299 117 - 22224390 ---ACUUUGGCCAAUCUCUGAAUGAGAUCGGAACAGUUGCUUCCCGAGGCAGAUUCAAUUGACUUCUUAAUGGCCAAAAAGGAUUCACUGGGCCUGAAGCCCUGAAUGAGUUUACACGGU ---..(((((((((((((.....)))))..(((...((((((....)))))).)))..............))))))))..(((((((..((((.....))))....)))))))....... ( -34.30) >DroSec_CAF1 3382 113 - 1 ---ACUUUGGCCAAUCUCUGAAUGAGAUGGGAACAGUUGCCUCCCGAGGUAGAUUCAAUUGAC----UAAUGGCCAAAAAGGAUUCACUGGACCUGAAGCCCUGAAUGAGUUUAUUCGGU ---..(((((((((((((.....)))))..(((...((((((....)))))).))).......----...))))))))..((.((((.......)))).))(((((((....))))))). ( -31.80) >DroSim_CAF1 3974 113 - 1 ---ACUUUGGCCAAUCUCUGAAUGAGAUCGGAACAGUUGCCUCCCGAGGCAGAUUCAAUUGAC----UAAUGGCCAAAAAGGAUUCACUGGGCCUGAAGCCCUGAAUAAGUUUAUCCGGU ---..(((((((((((((.....)))))..(((...((((((....)))))).))).......----...))))))))..((((..(((((((.....))))......)))..))))... ( -37.40) >DroYak_CAF1 4146 115 - 1 UCCACUUUGGCCAAUCUCUGAAUGCGAUCGGAUCAGCUUCUUCUCGAGGCAGAUCCAACUGAC----UAAUGGCCAAAAAGGUUUC-CUGGGCCUGAAGCCAUGAAUGAGUUCAUCCGGU .((..(((((((((((.(.....).))).(((((.(((((.....))))).))))).......----...))))))))..((((((-........))))))(((((....)))))..)). ( -35.80) >consensus ___ACUUUGGCCAAUCUCUGAAUGAGAUCGGAACAGUUGCCUCCCGAGGCAGAUUCAAUUGAC____UAAUGGCCAAAAAGGAUUCACUGGGCCUGAAGCCCUGAAUGAGUUUAUCCGGU .....(((((((((((((.....))))).(((....((((((....)))))).)))..............))))))))..(((((((..((((.....))))....)))))))....... (-27.89 = -28.70 + 0.81)



| Location | 20,883,339 – 20,883,454 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.63 |
| Mean single sequence MFE | -32.42 |
| Consensus MFE | -27.73 |
| Energy contribution | -27.48 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.603405 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20883339 115 - 22224390 GCAAUUUGCGAACGAUCGUCGGCCAGAGAUAAUGCUCU-----ACUUUGGCCAAUCUCUGAAUGAGAUCGGAACAGUUGCUUCCCGAGGCAGAUUCAAUUGACUUCUUAAUGGCCAAAAA ((((((......(((((.(((..(((((((...(((..-----.....)))..)))))))..))))))))....)))))).......((((((.((....))..))).....)))..... ( -29.50) >DroSec_CAF1 3422 111 - 1 GCAAUUUGCGAACGAUCGUCGGCCAGAGAUAAUGCUCU-----ACUUUGGCCAAUCUCUGAAUGAGAUGGGAACAGUUGCCUCCCGAGGUAGAUUCAAUUGAC----UAAUGGCCAAAAA .......((((....)))).(((((.((.((((..(((-----(((((((...(((((.....)))))(((........))).))))))))))....)))).)----)..)))))..... ( -32.00) >DroSim_CAF1 4014 111 - 1 GCAAUUUGCGAACGAUCGUCGGCCAGAGAUAAUGCUCU-----ACUUUGGCCAAUCUCUGAAUGAGAUCGGAACAGUUGCCUCCCGAGGCAGAUUCAAUUGAC----UAAUGGCCAAAAA ((.(((..(((.(((((.(((..(((((((...(((..-----.....)))..)))))))..))))))))(((...((((((....)))))).)))..)))..----.))).))...... ( -32.10) >DroYak_CAF1 4185 116 - 1 GCAAUUUGCGAACGAUCGUCGGCCAGAGAUAAUGCUCUGCUCCACUUUGGCCAAUCUCUGAAUGCGAUCGGAUCAGCUUCUUCUCGAGGCAGAUCCAACUGAC----UAAUGGCCAAAAA .......((((....)))).((.(((((......)))))..))..(((((((((((.(.....).))).(((((.(((((.....))))).))))).......----...)))))))).. ( -36.10) >consensus GCAAUUUGCGAACGAUCGUCGGCCAGAGAUAAUGCUCU_____ACUUUGGCCAAUCUCUGAAUGAGAUCGGAACAGUUGCCUCCCGAGGCAGAUUCAAUUGAC____UAAUGGCCAAAAA ((.....))(..((.....))..)((((......)))).......(((((((((((((.....))))).(((....((((((....)))))).)))..............)))))))).. (-27.73 = -27.48 + -0.25)


| Location | 20,883,379 – 20,883,494 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.67 |
| Mean single sequence MFE | -26.27 |
| Consensus MFE | -22.30 |
| Energy contribution | -22.12 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.652405 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20883379 115 - 22224390 AUAAAUAAAAAGUACACAUAAGUAUUAAAACCGAAACAGAGCAAUUUGCGAACGAUCGUCGGCCAGAGAUAAUGCUCU-----ACUUUGGCCAAUCUCUGAAUGAGAUCGGAACAGUUGC ..........(((((......)))))..............((((((......(((((.(((..(((((((...(((..-----.....)))..)))))))..))))))))....)))))) ( -26.20) >DroSec_CAF1 3458 108 - 1 -------AAAAGUACACAUAAGUAUUAAAACCGAAACGGAGCAAUUUGCGAACGAUCGUCGGCCAGAGAUAAUGCUCU-----ACUUUGGCCAAUCUCUGAAUGAGAUGGGAACAGUUGC -------...(((((......)))))....(((...))).((((((.((((....)))).((((((((.((......)-----))))))))).(((((.....)))))......)))))) ( -26.80) >DroSim_CAF1 4050 108 - 1 -------AAAAGUACACAUAAGUAUUAAAACCGAAACAGAGCAAUUUGCGAACGAUCGUCGGCCAGAGAUAAUGCUCU-----ACUUUGGCCAAUCUCUGAAUGAGAUCGGAACAGUUGC -------...(((((......)))))..............((((((......(((((.(((..(((((((...(((..-----.....)))..)))))))..))))))))....)))))) ( -26.20) >DroYak_CAF1 4221 113 - 1 -------AAAAGUACACAUAAGUAUUAAAACCAAAACAGAGCAAUUUGCGAACGAUCGUCGGCCAGAGAUAAUGCUCUGCUCCACUUUGGCCAAUCUCUGAAUGCGAUCGGAUCAGCUUC -------...(((((......)))))..........((((....)))).((.(((((((.((((((((.....((...))....))))))))..((...))..)))))))..))...... ( -25.90) >consensus _______AAAAGUACACAUAAGUAUUAAAACCGAAACAGAGCAAUUUGCGAACGAUCGUCGGCCAGAGAUAAUGCUCU_____ACUUUGGCCAAUCUCUGAAUGAGAUCGGAACAGUUGC ..........(((((......)))))..........(((((......((((....)))).((((((((................))))))))...))))).................... (-22.30 = -22.12 + -0.19)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:03:21 2006