
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,882,783 – 20,882,893 |
| Length | 110 |
| Max. P | 0.575353 |

| Location | 20,882,783 – 20,882,893 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.12 |
| Mean single sequence MFE | -29.80 |
| Consensus MFE | -20.05 |
| Energy contribution | -22.54 |
| Covariance contribution | 2.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.25 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.575353 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
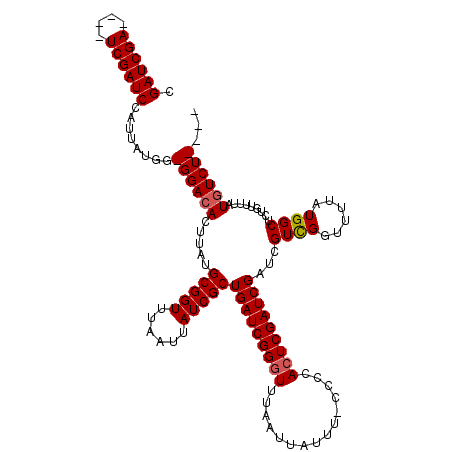
>X_DroMel_CAF1 20882783 110 + 22224390 ----AGACAUAAAACAGAGCCAUAAAACCAACGAUCGAUCGAGUGGGG-AAAUAAUUAAACCCGAUCAGCGAUAAUUAAACCGCAUAAGUGUCC-CCAUAAUGGAUCGA----UCGAUCG ----...........................(((((((((((((((((-(..((((((....((.....)).))))))...(((....))))))-))))......))))----))))))) ( -29.10) >DroSec_CAF1 2644 114 + 1 ----AGAUAUAAAACAGAGCCAUAAAACCGACGAUCGAUCGAUUGGGG-AAAUAAUUAAACCCGAUCAGCGAUAAUUAAACCGCAUAAGUGUCC-CCAUAAUGGAUCGAUCGAUCGAUCG ----........................(((((((((((((((((((.-...........))))))).(((..........)))......((((-.......)))).))))))))).))) ( -32.10) >DroSim_CAF1 3445 114 + 1 ----AGACAUAAAACAGAGCCAUAAAACCGACGAUCGAUCGAGUGGGG-AAAUAAUUAAACCCGAUCAGCGAUAAUUAAACCGCAUAAGUGUCC-CCAUAAUGGAUCGAUCGAUCGAUCG ----........................((((((((((((((((((((-(..((((((....((.....)).))))))...(((....))))))-))))......))))))))))).))) ( -32.10) >DroYak_CAF1 3580 112 + 1 AAUAAGACAUAAAACAGAGCCAUAAAAUCAACGAUCGAUCGAGUGGGGCAACUAAUUAAACCCGAUCAGCGAUAAUUAAACCGCAUAAGUGUCCCCCAUAAUGGAUC--------GAUCG ...............................(((((((((.(((((((..(((.(((......)))..(((..........)))...)))...))))))..).))))--------))))) ( -25.90) >consensus ____AGACAUAAAACAGAGCCAUAAAACCAACGAUCGAUCGAGUGGGG_AAAUAAUUAAACCCGAUCAGCGAUAAUUAAACCGCAUAAGUGUCC_CCAUAAUGGAUCGA____UCGAUCG ...............................(((((((((((..(((.............)))((((.(((..........)))....(((.....)))....))))..))))))))))) (-20.05 = -22.54 + 2.50)



| Location | 20,882,783 – 20,882,893 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.12 |
| Mean single sequence MFE | -31.88 |
| Consensus MFE | -21.57 |
| Energy contribution | -22.14 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.68 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.517193 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20882783 110 - 22224390 CGAUCGA----UCGAUCCAUUAUGG-GGACACUUAUGCGGUUUAAUUAUCGCUGAUCGGGUUUAAUUAUUU-CCCCACUCGAUCGAUCGUUGGUUUUAUGGCUCUGUUUUAUGUCU---- (((((((----((((((.....((.-...)).....(((((......))))).))))(((...........-.)))....)))))))))..........(((..........))).---- ( -29.80) >DroSec_CAF1 2644 114 - 1 CGAUCGAUCGAUCGAUCCAUUAUGG-GGACACUUAUGCGGUUUAAUUAUCGCUGAUCGGGUUUAAUUAUUU-CCCCAAUCGAUCGAUCGUCGGUUUUAUGGCUCUGUUUUAUAUCU---- (((.((((((((((((......(((-(((.......(((((......)))))(((......)))......)-))))))))))))))))))))........................---- ( -35.30) >DroSim_CAF1 3445 114 - 1 CGAUCGAUCGAUCGAUCCAUUAUGG-GGACACUUAUGCGGUUUAAUUAUCGCUGAUCGGGUUUAAUUAUUU-CCCCACUCGAUCGAUCGUCGGUUUUAUGGCUCUGUUUUAUGUCU---- .((((((.(((((((((.....(((-(((.......(((((......)))))(((......)))......)-)))))...)))))))))))))))....(((..........))).---- ( -34.30) >DroYak_CAF1 3580 112 - 1 CGAUC--------GAUCCAUUAUGGGGGACACUUAUGCGGUUUAAUUAUCGCUGAUCGGGUUUAAUUAGUUGCCCCACUCGAUCGAUCGUUGAUUUUAUGGCUCUGUUUUAUGUCUUAUU (((((--------((((.....((((((((......(((((......)))))(((......)))....))).)))))...)))))))))..........(((..........)))..... ( -28.10) >consensus CGAUCGA____UCGAUCCAUUAUGG_GGACACUUAUGCGGUUUAAUUAUCGCUGAUCGGGUUUAAUUAUUU_CCCCACUCGAUCGAUCGUCGGUUUUAUGGCUCUGUUUUAUGUCU____ .((((((....))))))........((((((.....(((((......)))))(((((((((...............)))))))))...((((......)))).........))))))... (-21.57 = -22.14 + 0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:03:17 2006