
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,882,360 – 20,882,598 |
| Length | 238 |
| Max. P | 0.875033 |

| Location | 20,882,360 – 20,882,478 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.10 |
| Mean single sequence MFE | -35.11 |
| Consensus MFE | -22.35 |
| Energy contribution | -23.73 |
| Covariance contribution | 1.37 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.875033 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20882360 118 + 22224390 AAAGUGGCGAAAAACUUCUAGAGAGUGGGAGAGAAAACUCCGCUGCAGUUCAGUCGG--UUGUUAUCAGCAGCAGUUGCUUGUUGACGACAAUCGAUUCUGAUUCUGAUUCUGCUUCCAA ..(((((.(.....((((((.....))))))......).)))))((((.((((((((--((((..((((((((....)).))))))..))))))))........))))..))))...... ( -38.30) >DroSec_CAF1 2246 100 + 1 AAAGGGGCGGAAAACUUCCAGAGAGUGGGAGAGAAAACUCCG--------------------UUAUCAGCAGCAGUUGCUUGUUGACGGCAAUCGAUUCAGAUUCUGAUUCUGAUUCCAA ...((((.......((((((.....))))))......))))(--------------------(..((((((((....)).))))))..))....((.(((((.......))))).))... ( -29.32) >DroSim_CAF1 3034 117 + 1 AGAGGGGCGGAAAACUUCCAGAGAGUGGGAGAGAAAACUCCGCUGCAGUUUAGUCGG---UGUUAUCAGCAGCAGUUGCUUGUUGACGGCAAUCGAUUCAGAUUCUGAUUCUGAUUCCAA ...(.((((((...((((((.....)))))).......)))))).)......(((((---(((..((((((((....)).))))))..)).))))))(((((.......)))))...... ( -38.40) >DroYak_CAF1 3196 105 + 1 AAAGUGGCGGAAAACUUCCAGAGAGUGGGAGAUGGAACUCCGCUGCAGUUUGGUCAGUGUUGUUAUCAGCAGCAGUUGCUUGUUGACGGCAAUCGAUUUUGAUUU--------------- ...(..(((((...((((((.....)))))).......)))))..).....((((((.(((((..((((((((....)).))))))..))))).....)))))).--------------- ( -34.40) >consensus AAAGGGGCGGAAAACUUCCAGAGAGUGGGAGAGAAAACUCCGCUGCAGUUUAGUCGG___UGUUAUCAGCAGCAGUUGCUUGUUGACGGCAAUCGAUUCAGAUUCUGAUUCUGAUUCCAA ...((((((((...((((((.....)))))).......))))))))...............((..((((((((....)).))))))..))((((......))))................ (-22.35 = -23.73 + 1.37)



| Location | 20,882,438 – 20,882,558 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.67 |
| Mean single sequence MFE | -24.45 |
| Consensus MFE | -16.88 |
| Energy contribution | -17.62 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.69 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20882438 120 - 22224390 AACAAAAACAAAAAAGCAAACAAAUGUCGAUGGUUGCACACGACUGCGUUGUGUGAACAAUGGAGAGCUGGUUAACAGUAUUGGAAGCAGAAUCAGAAUCAGAAUCGAUUGUCGUCAACA ........................(((.((((....((((((((...)))))))).(((((.((...((((((....((.(((....))).))...))))))..)).))))))))).))) ( -27.60) >DroSec_CAF1 2306 117 - 1 AACAAAAA-AAAAAAGCAAACAAAUGUCGAUCGCUGCACACGACGGCGUUGUGUGAACAAUUG--AGCUGGUAAACAGUAUUGGAAUCAGAAUCAGAAUCUGAAUCGAUUGCCGUCAACA ........-...............(((.(((.((..((((((((...))))))))...(((((--(((((.....)))).......(((((.......))))).)))))))).))).))) ( -26.40) >DroSim_CAF1 3111 117 - 1 AACAAAAA-AAAAACGCAAACAAAUGUCGAUCGCUGCACACGACGGCGUUGUGUGAACAAUGG--AGCUGGUAAACAGUAUUGGAAUCAGAAUCAGAAUCUGAAUCGAUUGCCGUCAACA ........-.....((((....(((((((.(((.......)))))))))).))))....((((--.((((.....))))....((.(((((.......))))).)).....))))..... ( -26.10) >DroYak_CAF1 3276 99 - 1 AAAAACAA-AUAAACGCUAACAAAUGUCGAUCGCUGCACACGACGGCGUUGUGUGAACAAUGG--AGCUGGUAAACA------------------AAAUCAAAAUCGAUUGCCGUCAACA ........-...............(((.(((.((..((((((((...))))))))...(((.(--(..((((.....------------------..))))...)).))))).))).))) ( -17.70) >consensus AACAAAAA_AAAAAAGCAAACAAAUGUCGAUCGCUGCACACGACGGCGUUGUGUGAACAAUGG__AGCUGGUAAACAGUAUUGGAAUCAGAAUCAGAAUCAGAAUCGAUUGCCGUCAACA .......................(((.((((((...((((((((...))))))))...........((((.....))))..........................)))))).)))..... (-16.88 = -17.62 + 0.75)

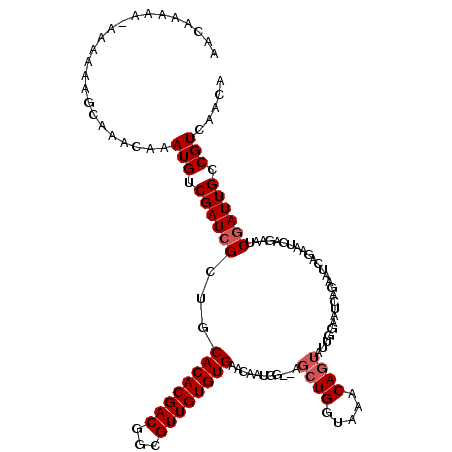
| Location | 20,882,478 – 20,882,598 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.22 |
| Mean single sequence MFE | -29.34 |
| Consensus MFE | -23.14 |
| Energy contribution | -23.08 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.79 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.523093 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20882478 120 - 22224390 GCAGAAGUAACGGGCAGUCGGCAACAGUGGGCCGUUCGAAAACAAAAACAAAAAAGCAAACAAAUGUCGAUGGUUGCACACGACUGCGUUGUGUGAACAAUGGAGAGCUGGUUAACAGUA ((((..((..(((((.(((.((....)).))).))))).................((((.((........)).))))..))..))))(((((....))))).....((((.....)))). ( -30.00) >DroSec_CAF1 2346 115 - 1 GCAGAAGUAACGGGCAGUCGGCAACAGUGGGCAGUUG--AAACAAAAA-AAAAAAGCAAACAAAUGUCGAUCGCUGCACACGACGGCGUUGUGUGAACAAUUG--AGCUGGUAAACAGUA .(((..((....(((.(((((((...((..((..((.--.........-..))..))..))...))))))).))).((((((((...)))))))).))..)))--.((((.....)))). ( -28.20) >DroSim_CAF1 3151 115 - 1 GCAGAAGUAACGGGCAGUCGGCAACAGUGGGCAGUUG--AAACAAAAA-AAAAACGCAAACAAAUGUCGAUCGCUGCACACGACGGCGUUGUGUGAACAAUGG--AGCUGGUAAACAGUA ............(((.(((((((...((..((.(((.--.........-...)))))..))...))))))).))).((((((((...))))))))........--.((((.....)))). ( -29.62) >DroYak_CAF1 3301 110 - 1 GCAGAAGUAACGGCCGGUCGGCAACAGUGGGUCGU----AAAAAACAA-AUAAACGCUAACAAAUGUCGAUCGCUGCACACGACGGCGUUGUGUGAACAAUGG--AGCUGGUAAACA--- .(((......((((.((((((((...((.((.(((----.........-....))))).))...))))))))))))((((((((...))))))))........--..))).......--- ( -29.52) >consensus GCAGAAGUAACGGGCAGUCGGCAACAGUGGGCAGUUG__AAACAAAAA_AAAAAAGCAAACAAAUGUCGAUCGCUGCACACGACGGCGUUGUGUGAACAAUGG__AGCUGGUAAACAGUA .(((........(((.(((((((...((..((.......................))..))...))))))).))).((((((((...))))))))............))).......... (-23.14 = -23.08 + -0.06)

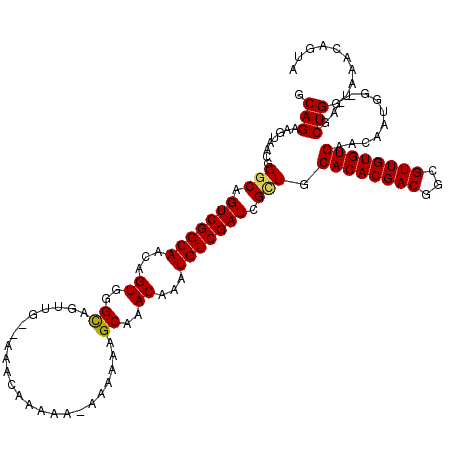

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:03:15 2006