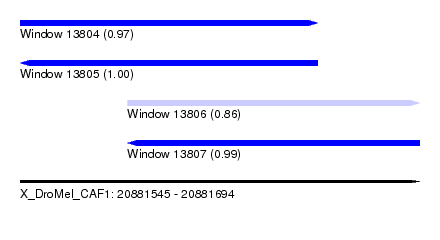
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,881,545 – 20,881,694 |
| Length | 149 |
| Max. P | 0.997324 |

| Location | 20,881,545 – 20,881,656 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.43 |
| Mean single sequence MFE | -40.02 |
| Consensus MFE | -31.21 |
| Energy contribution | -32.27 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.974732 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
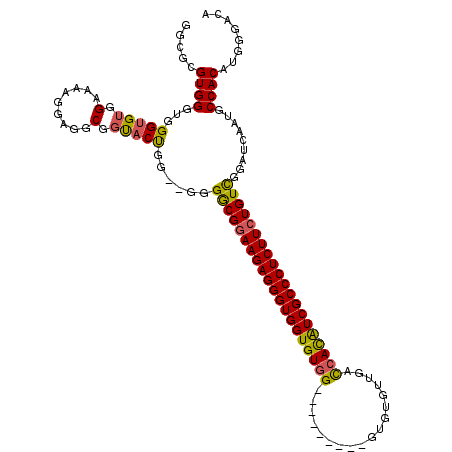
>X_DroMel_CAF1 20881545 111 + 22224390 ACUUGGAGAGCCGUCGAAAGACAUAUGCGUCAGCAUCAGCUGUCCAAUGUGGCAAUCAUUCGACAGAAGAGGGCGAUGUGGCCAACACAC---------CCACACCACCCUCUUUCGUCU (((((((.(((.(((....)))..((((....))))..))).))))).))...........(((..(((((((.(.(((((.........---------))))).).)))))))..))). ( -44.80) >DroSec_CAF1 1495 111 + 1 ACUUGGAGAGCCGUCGAACGACAUCUGCGUCAGCAUCAGCUGUCCCAUGUGGCAUUGAUCCGACAGAAGAGGGCGAUGUGGUCAACACAC---------CCACACCACCCUCUUCCGCCC ..(((((..((((((....)))...((.(.((((....)))).).))...))).....)))))..((((((((.(.(((((.........---------))))).).))))))))..... ( -39.10) >DroSim_CAF1 2292 111 + 1 ACUUGGAGAGCCGUCGAACGACAUCUGCGUCAGCAUCAGCUGUCCCAUGUGGCAUUGAUCCGACAGAAGAGGGCGAUGUGGUCAACACAC---------CCACACCACCCUCUUCCGCCC ..(((((..((((((....)))...((.(.((((....)))).).))...))).....)))))..((((((((.(.(((((.........---------))))).).))))))))..... ( -39.10) >DroYak_CAF1 2389 120 + 1 ACAUGGAGUGUCGCUGAAAGACAUCUGCGUCAGCGUCUGCUGUCCAAUGUGGCAUUGAUCCAACAGAAGAGGGCGACAUGAUCAACACCACGACAUGGUCAACACCACCCUCUUCCGCCC ((((((((((.((((((.((....))...)))))).).))..))).))))(((..((......))((((((((.(...((((((...........))))))....).)))))))).))). ( -37.10) >consensus ACUUGGAGAGCCGUCGAAAGACAUCUGCGUCAGCAUCAGCUGUCCAAUGUGGCAUUGAUCCGACAGAAGAGGGCGAUGUGGUCAACACAC_________CCACACCACCCUCUUCCGCCC ..(((((..((((((....)))......(.((((....)))).)......))).....)))))..((((((((.(.(((((..................))))).).))))))))..... (-31.21 = -32.27 + 1.06)


| Location | 20,881,545 – 20,881,656 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.43 |
| Mean single sequence MFE | -51.65 |
| Consensus MFE | -42.37 |
| Energy contribution | -42.49 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.18 |
| Mean z-score | -3.59 |
| Structure conservation index | 0.82 |
| SVM decision value | 2.84 |
| SVM RNA-class probability | 0.997324 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20881545 111 - 22224390 AGACGAAAGAGGGUGGUGUGG---------GUGUGUUGGCCACAUCGCCCUCUUCUGUCGAAUGAUUGCCACAUUGGACAGCUGAUGCUGACGCAUAUGUCUUUCGACGGCUCUCCAAGU .((((.(((((((((((((((---------.(.....).))))))))))))))).))))..............(((((.((((.((((....))))..(((....))))))).))))).. ( -49.10) >DroSec_CAF1 1495 111 - 1 GGGCGGAAGAGGGUGGUGUGG---------GUGUGUUGACCACAUCGCCCUCUUCUGUCGGAUCAAUGCCACAUGGGACAGCUGAUGCUGACGCAGAUGUCGUUCGACGGCUCUCCAAGU .((((((((((((((((((((---------.........))))))))))))))))))))(((.....(((...((.(.((((....)))).).))...(((....))))))..))).... ( -56.60) >DroSim_CAF1 2292 111 - 1 GGGCGGAAGAGGGUGGUGUGG---------GUGUGUUGACCACAUCGCCCUCUUCUGUCGGAUCAAUGCCACAUGGGACAGCUGAUGCUGACGCAGAUGUCGUUCGACGGCUCUCCAAGU .((((((((((((((((((((---------.........))))))))))))))))))))(((.....(((...((.(.((((....)))).).))...(((....))))))..))).... ( -56.60) >DroYak_CAF1 2389 120 - 1 GGGCGGAAGAGGGUGGUGUUGACCAUGUCGUGGUGUUGAUCAUGUCGCCCUCUUCUGUUGGAUCAAUGCCACAUUGGACAGCAGACGCUGACGCAGAUGUCUUUCAGCGACACUCCAUGU .((((((((((((((((....)))..(.((((((....)))))).))))))))))))))(((((((((...)))))).((((....)))).(((.((......)).)))....))).... ( -44.30) >consensus GGGCGGAAGAGGGUGGUGUGG_________GUGUGUUGACCACAUCGCCCUCUUCUGUCGGAUCAAUGCCACAUGGGACAGCUGAUGCUGACGCAGAUGUCGUUCGACGGCUCUCCAAGU .((((((((((((((((((((..................))))))))))))))))))))(((.....(((........((((....))))........(((....))))))..))).... (-42.37 = -42.49 + 0.13)



| Location | 20,881,585 – 20,881,694 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.34 |
| Mean single sequence MFE | -28.73 |
| Consensus MFE | -20.27 |
| Energy contribution | -21.65 |
| Covariance contribution | 1.38 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.860762 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20881585 109 + 22224390 UGUCCAAUGUGGCAAUCAUUCGACAGAAGAGGGCGAUGUGGCCAACACAC---------CCACACCACCCUCUUUCGUCUC--ACAGUACCGCCUCCUUUUCCACACCCACCCACGCGCC .......(((((.........(((..(((((((.(.(((((.........---------))))).).)))))))..)))..--...(.....)........))))).............. ( -27.40) >DroSec_CAF1 1535 109 + 1 UGUCCCAUGUGGCAUUGAUCCGACAGAAGAGGGCGAUGUGGUCAACACAC---------CCACACCACCCUCUUCCGCCCC--CCAGUACCGCCUCCUUUUCCCCACCCACCCACGCGCC ........((((.((((....(.(.((((((((.(.(((((.........---------))))).).)))))))).))...--.)))).))))........................... ( -28.00) >DroSim_CAF1 2332 110 + 1 UGUCCCAUGUGGCAUUGAUCCGACAGAAGAGGGCGAUGUGGUCAACACAC---------CCACACCACCCUCUUCCGCCCCC-CCGGUACCGCCUCCUUUUCCACACCCACCCACGCGCC .......(((((....((..((...((((((((.(.(((((.........---------))))).).)))))))).(((...-..)))..))..)).....))))).............. ( -29.80) >DroYak_CAF1 2429 119 + 1 UGUCCAAUGUGGCAUUGAUCCAACAGAAGAGGGCGACAUGAUCAACACCACGACAUGGUCAACACCACCCUCUUCCGCCCCCCCCAGAGCCGCC-CCUUUGCCACACCCACCCACGCGCC .......(((((((..(........((((((((.(...((((((...........))))))....).)))))))).((.(......).))....-.)..))))))).............. ( -29.70) >consensus UGUCCAAUGUGGCAUUGAUCCGACAGAAGAGGGCGAUGUGGUCAACACAC_________CCACACCACCCUCUUCCGCCCC__CCAGUACCGCCUCCUUUUCCACACCCACCCACGCGCC ........((((.((((........((((((((.(.(((((..................))))).).)))))))).........)))).))))........................... (-20.27 = -21.65 + 1.38)

| Location | 20,881,585 – 20,881,694 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.34 |
| Mean single sequence MFE | -47.65 |
| Consensus MFE | -38.86 |
| Energy contribution | -38.55 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.23 |
| Mean z-score | -3.06 |
| Structure conservation index | 0.82 |
| SVM decision value | 2.48 |
| SVM RNA-class probability | 0.994483 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20881585 109 - 22224390 GGCGCGUGGGUGGGUGUGGAAAAGGAGGCGGUACUGU--GAGACGAAAGAGGGUGGUGUGG---------GUGUGUUGGCCACAUCGCCCUCUUCUGUCGAAUGAUUGCCACAUUGGACA .((((.(....).)))).........((((((...((--..((((.(((((((((((((((---------.(.....).))))))))))))))).))))..)).)))))).......... ( -44.40) >DroSec_CAF1 1535 109 - 1 GGCGCGUGGGUGGGUGGGGAAAAGGAGGCGGUACUGG--GGGGCGGAAGAGGGUGGUGUGG---------GUGUGUUGACCACAUCGCCCUCUUCUGUCGGAUCAAUGCCACAUGGGACA ..(.((((....((((..((.....((......))..--..((((((((((((((((((((---------.........))))))))))))))))))))...))..)))).)))).)... ( -48.80) >DroSim_CAF1 2332 110 - 1 GGCGCGUGGGUGGGUGUGGAAAAGGAGGCGGUACCGG-GGGGGCGGAAGAGGGUGGUGUGG---------GUGUGUUGACCACAUCGCCCUCUUCUGUCGGAUCAAUGCCACAUGGGACA ..(.((((....(((((.((........((....)).-...((((((((((((((((((((---------.........))))))))))))))))))))...)).))))).)))).)... ( -50.40) >DroYak_CAF1 2429 119 - 1 GGCGCGUGGGUGGGUGUGGCAAAGG-GGCGGCUCUGGGGGGGGCGGAAGAGGGUGGUGUUGACCAUGUCGUGGUGUUGAUCAUGUCGCCCUCUUCUGUUGGAUCAAUGCCACAUUGGACA ..(((....)))(((((((((..((-..((((...(((((((((((......(((((....)))))..((((((....))))))))))))))))).))))..))..)))))))))..... ( -47.00) >consensus GGCGCGUGGGUGGGUGUGGAAAAGGAGGCGGUACUGG__GGGGCGGAAGAGGGUGGUGUGG_________GUGUGUUGACCACAUCGCCCUCUUCUGUCGGAUCAAUGCCACAUGGGACA .....((((...(((((.(.........).)))))......((((((((((((((((((((..................)))))))))))))))))))).........))))........ (-38.86 = -38.55 + -0.31)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:03:11 2006