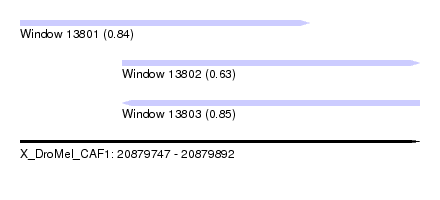
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,879,747 – 20,879,892 |
| Length | 145 |
| Max. P | 0.848977 |

| Location | 20,879,747 – 20,879,852 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 79.20 |
| Mean single sequence MFE | -35.05 |
| Consensus MFE | -24.03 |
| Energy contribution | -24.37 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.837598 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20879747 105 + 22224390 AAGUAGCCGCAGCCCAAUUCGAAAUGCUAAUGAUAACGAAAAUCUGGUUGGGUGAAUCACCCAGUGAUUCAGUGGGGCUUGAC------GAGGAUCGCCUCCUAGCCAGAU ......((((.(((((((..((....................))..)))))))(((((((...))))))).))))((((....------((((....))))..)))).... ( -33.25) >DroSec_CAF1 7325 101 + 1 ----AGCCGCAACCCAAUUCGAAAUGCUAAUGAUAACGAAUGUCUGGUUGGGUGAAUCACCCGGUGAUUCAGUGGGGCUCGGC------GAGGAUCGCCUCCUAGCCAGAU ----..((((.(((((((..((....(..........)....))..)))))))(((((((...))))))).))))((((.((.------((((....)))))))))).... ( -35.80) >DroEre_CAF1 7397 111 + 1 AAGUAACAUUAGACCGACACGAAACAGUAAUGAUAAGGAAAGUCUGGGUGGGUGAAUCACCCAGUGAUUCAGUGGGGCUUGGCUCCACACAGGAUCCACCCCUGGCCAGAU ........................((....))....((..((...(((((((((((((((...))))))).(((((((...))))))).....))))))))))..)).... ( -36.10) >consensus AAGUAGCCGCAGCCCAAUUCGAAAUGCUAAUGAUAACGAAAGUCUGGUUGGGUGAAUCACCCAGUGAUUCAGUGGGGCUUGGC______GAGGAUCGCCUCCUAGCCAGAU .........................................((((((((((.((((((((...))))))))..(((((..................))))))))))))))) (-24.03 = -24.37 + 0.34)

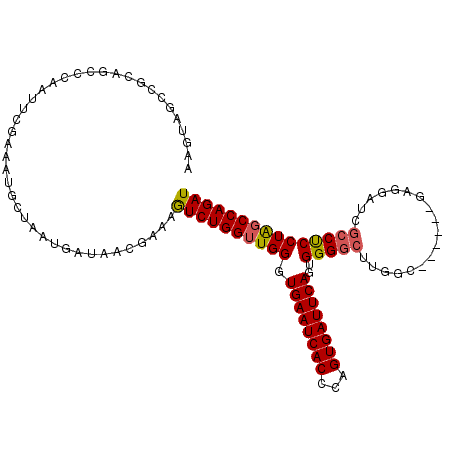

| Location | 20,879,784 – 20,879,892 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 84.52 |
| Mean single sequence MFE | -38.90 |
| Consensus MFE | -28.09 |
| Energy contribution | -27.76 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.633283 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20879784 108 + 22224390 GAAAAUCUGGUUGGGUGAAUCACCCAGUGAUUCAGUGGGGCUUGAC------GAGGAUCGCCUCCUAGCCAGAUCAGUUAAAAGUCCCCCACUGAAACUGACCACUUCUUGCGA .......((((((((((...)))))(((..((((((((((...(((------((((....)))).((((.......))))...))))))))))))))))))))).......... ( -37.60) >DroSec_CAF1 7358 108 + 1 GAAUGUCUGGUUGGGUGAAUCACCCGGUGAUUCAGUGGGGCUCGGC------GAGGAUCGCCUCCUAGCCAGAUCUGAUUAAAGUCCCCCACUGAAACUGACCACUUCUUGUGA .......((((((((((...)))))(((..((((((((((...(((------.((((.....)))).))).((.((......)))))))))))))))))))))).......... ( -40.50) >DroEre_CAF1 7434 113 + 1 GAAAGUCUGGGUGGGUGAAUCACCCAGUGAUUCAGUGGGGCUUGGCUCCACACAGGAUCCACCCCUGGCCAGAUCAGAUUAAAGUCC-CCACAGAACCUGACCAUUUCUUGCGG (.(((..(((..(((((((((((...))))))).(((((((((((.(((.....))).))....((((.....))))....))).))-))))...))))..)))...))).).. ( -38.60) >consensus GAAAGUCUGGUUGGGUGAAUCACCCAGUGAUUCAGUGGGGCUUGGC______GAGGAUCGCCUCCUAGCCAGAUCAGAUUAAAGUCCCCCACUGAAACUGACCACUUCUUGCGA (((.(((.(((((((((...))))))....((((((((((...(((.........((((..((...))...))))........)))))))))))))))))))...)))...... (-28.09 = -27.76 + -0.33)


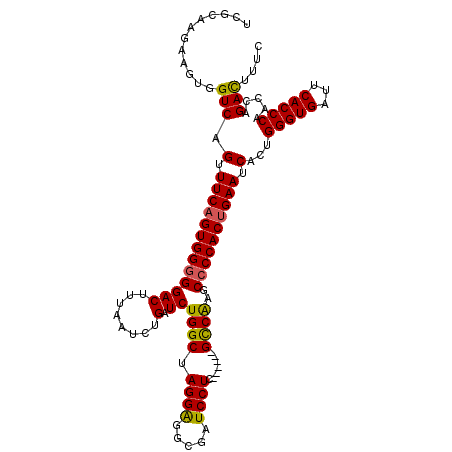
| Location | 20,879,784 – 20,879,892 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 84.52 |
| Mean single sequence MFE | -41.17 |
| Consensus MFE | -28.98 |
| Energy contribution | -28.77 |
| Covariance contribution | -0.21 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.848977 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20879784 108 - 22224390 UCGCAAGAAGUGGUCAGUUUCAGUGGGGGACUUUUAACUGAUCUGGCUAGGAGGCGAUCCUC------GUCAAGCCCCACUGAAUCACUGGGUGAUUCACCCAACCAGAUUUUC ((....))..((((..(.(((((((((((((........).))((((.((((.....)))).------))))..)))))))))).)..((((((...))))))))))....... ( -40.00) >DroSec_CAF1 7358 108 - 1 UCACAAGAAGUGGUCAGUUUCAGUGGGGGACUUUAAUCAGAUCUGGCUAGGAGGCGAUCCUC------GCCGAGCCCCACUGAAUCACCGGGUGAUUCACCCAACCAGACAUUC .(((.....)))(((.(.((((((((((((......)).....((((.((((.....)))).------))))..)))))))))).)...(((((...))))).....))).... ( -41.40) >DroEre_CAF1 7434 113 - 1 CCGCAAGAAAUGGUCAGGUUCUGUGG-GGACUUUAAUCUGAUCUGGCCAGGGGUGGAUCCUGUGUGGAGCCAAGCCCCACUGAAUCACUGGGUGAUUCACCCACCCAGACUUUC .(....)...((((((((((....(.-...)........))))))))))(((((((.(((.....))).))..))))).........(((((((.......)))))))...... ( -42.10) >consensus UCGCAAGAAGUGGUCAGUUUCAGUGGGGGACUUUAAUCUGAUCUGGCUAGGAGGCGAUCCUC______GCCAAGCCCCACUGAAUCACUGGGUGAUUCACCCAACCAGACUUUC ............(((.(.(((((((((((((........).))((((.((((.....)))).......))))..)))))))))).)...(((((...))))).....))).... (-28.98 = -28.77 + -0.21)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:03:08 2006