| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 20,871,974 – 20,872,090 |
| Length | 116 |
| Max. P | 0.641770 |

| Location | 20,871,974 – 20,872,090 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.28 |
| Mean single sequence MFE | -44.03 |
| Consensus MFE | -27.88 |
| Energy contribution | -29.97 |
| Covariance contribution | 2.09 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.641770 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 20871974 116 + 22224390 ----GCCACAUCACCUGUACUUUCAGCUCGGCUGGGCCUUUCUCUUCCUAGCCUUUUUGGCUCCACACGGUCCCUACGGCGCCCUGUGGAUGCGUGCCAUGCUGCUCAUUGGCUGCCUGA ----((((......(((......)))...(((((((..........)))))))....))))((((((.(((.((...)).))).)))))).(((.((((..........))))))).... ( -38.10) >DroVir_CAF1 279 120 + 1 AGCAGCCGCAUCAUCUGUACUUUCAGCUGGGCUGGGCCUUUCUUUUUCUGGCCUUUUUGGCGCCGCACGGUCCAUUUGGUUCGCUUUGGAUGCGCGCCACCCUGCUCAUUGGGUGCAUCA .(((.(((((.....)))......(((.(((..(((((...........)))))...((((((.(((...((((...((....)).)))))))))))))))).)))....)).))).... ( -42.30) >DroPse_CAF1 300 120 + 1 AGCAGCCGCACCACCUCUACUUUCAGCUCGGCUGGGCCUUCCUCUUUCUGGCCUUCCUGGCCCCCCACGGCCCCUUCGGCUCGCUGUGGAUGCGGGCGAUGCUGCUGAUCGGCUGCAUCA .(((((((..............(((((.(((..(((((...........)))))..)))((((((((((((..(....)...)))))))..).))))......))))).))))))).... ( -50.46) >DroGri_CAF1 309 120 + 1 AGCAGCCGCAUCAUCUCUACUUUCAGCUGGGCUGGGCCUUUCUCUUUCUGGCGUUUGUGGCGCCACACGGUCCGUUUGGGUCGCUAUGGAUGCGCGCCACACUGCUCAUCGGCUGCAUCA .(((((((.................((..((..(((.....)))..))..))((.((((((((.....(..((....))..)((.......))))))))))..))....))))))).... ( -46.10) >DroMoj_CAF1 300 120 + 1 AACAGCCGCAUCAUCUCUAUUUUCAGCUGGGCUGGGCCUUCCUCUUUCUGGCGUUUCUAGCGCCCCAUGGACCCUUCGGUUCGCUGUGGAUGCGCGCUACUAUGCUCAUCGGCUGCAUCA ..((((((((((...........((((.((((((((...(((.......(((((.....)))))....)))...))))))))))))..)))))(.((......)).)...)))))..... ( -39.92) >DroAna_CAF1 216 120 + 1 AGCAGCCGCACCAACUCUAUUUCCAACUGGGCUGGGCCUUCCUCUUCCUGGCCUUCCUGGCGCCGCACGGCCCCUUCGGCUCUCUGUGGAUGCGGGCCAUGCUCCUCAUCGGCUGCCUGA .(((((((..................(..((..(((((...........))))).))..)....(((.((((((.((.((.....)).)).).))))).))).......))))))).... ( -47.30) >consensus AGCAGCCGCAUCAUCUCUACUUUCAGCUGGGCUGGGCCUUCCUCUUUCUGGCCUUUCUGGCGCCACACGGUCCCUUCGGCUCGCUGUGGAUGCGCGCCACGCUGCUCAUCGGCUGCAUCA .(((((((((((.............((((((..(((((...........))))).))))))....(((((.((....))....))))))))))(((......))).....)))))).... (-27.88 = -29.97 + 2.09)

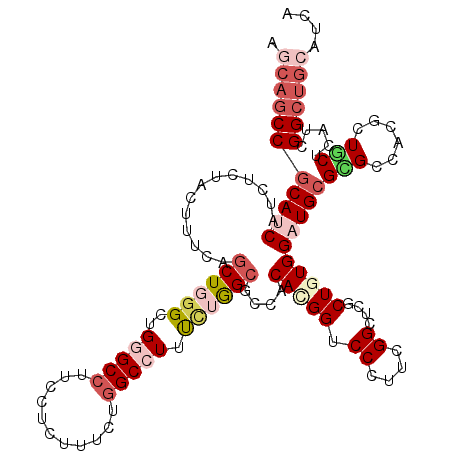

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:03:04 2006